Association of polymorphism rs10841855 in the glycogen synthase 2 gene with hypercholesterolemia and type 2 diabetes mellitus
Aннотация
Background: Glycogen synthase 2 (GYS2) catalyzes a key step in glycogenesis in the liver. Loss-of-function mutations in the GYS2 gene are associated with type 0a glycogenosis with characteristic fasting hypoglycemia and postprandial hyperglycemia. The association of the single nucleotide polymorphism rs10841855 (G>T) with the risk of type 2 diabetes mellitus (T2D) was first established in the European population, but data on the effect of this variant on the predisposition to T2D in the inhabitants of the Slavic ethnic group of Central Russia are not available in the literature. The aim of the study: To investigate the association of polymorphism rs10841855 (G>T) GYS2 with the risk of T2D in residents of Central Russia. Materials and methods: The study included 2668 unrelated individuals of Slavic origin, including 1387 patients with T2D and 1281 healthy volunteers. Genotyping of the rs10841855 polymorphism of the GYS2 gene was performed by the MassArray Analyzer-4 genetic analyzer. SNPStats software was used for statistical analysis of the data. Results: Linear regression established an association of the alternative rs10841855-T allele (OR 1.25; 95% CI 1.09-1.43; P=0.001) and the rs10841855-G/T genotype (OR 1.37; 95% CI 1, 13-1.66; P=0.0064) of GYS2 with an increased risk of T2D. The sex- and BMI-stratified analysis showed that the association of the rs10841855-G/T genotype (OR 1.71; 95% CI 1.32-2.22; P=0.0001) occurred only in females with BMI>25 kg/m2. In addition, carriage of the rs10841855-T allele was associated with a higher level of total cholesterol in the blood plasma of T2D patients (P=0.0068). According to the Roadmap Epigenomic Consortium, the rs10841855-T allele is associated with H3K4 histone methylation in the enhancer and promoter regions of the GYS2 gene in the liver. Conclusion: The association of rs10841855 of the GYS2 gene with an increased risk of T2D in residents of Central Russia was validated for the first time. The association may be explained by the low transcriptional activity of the glycogen synthase 2 gene in the carriers of the alternative allele of this polymorphism.
Ключевые слова: type 2 diabetes mellitus, single nucleotide polymorphism, GYS2, glycogen synthase, genetic predisposition
К сожалению, текст статьи доступен только на Английском
Introduction. According to the International Diabetes Federation, there are more than 537 million diabetic patients worldwide aged 20 to 79 years, with 90 percent having type 2 diabetes mellitus (T2D) [1]. The pathogenetic basis of the disease is dysfunction of pancreatic beta cells that secrete insulin, as well as insulin resistance of the liver, skeletal muscle, and adipose tissue, resulting in chronic hyperglycemia and disruption of all types of metabolism [2]. Type 2 diabetes complications include diabetic retinopathy, nephropathy, neuropathy, and macrovascular complications, which killed 6.7 million people in 2021 [1].
Type 2 diabetes mellitus is a classic multifactorial disease caused by genetic and environmental risk factors such as an excessive fat and refined carbohydrate diet [3, 4], sedentary lifestyle [5, 6], overweight and obesity [7, 8], chronic psycho-emotional stress [7, 8], and others. The genetic basis of the disease is being studied extensively: from 2007 to the present, 197 genome-wide association studies (GWAS) for T2D have been conducted worldwide, yielding a total of over four thousand single nucleotide markers (SNPs) [GWAS Catalog https://www.ebi.ac.uk/gwas/]. The majority of the detected loci are in non-coding regions of the genome, pseudogenes, and intergenic regions, which significantly complicates biological interpretation of the existing data layer and impedes clinical application of the results obtained. One of the polymorphic variants identified by R.A. Scott et al. in a genome-wide study in a European population of 159,208 people [9] became the SNP rs10841855 in the intron of the GYS2 gene, which encodes hepatic glycogen synthase 2. The enzyme catalyzes a critical step in hepatic glycogenesis, and its functional deficiency caused by GYS2 gene loss-of-function mutations is known as type 0a glycogenosis, which is characterized by fasting ketotic hypoglycemia and postprandial hyperglycemia due to the liver's inability to store glycogen [10, 11]. It should be noted that the liver plays a critical role in maintaining normal glucose homeostasis by catalyzing the conversion of glucose into glycogen under hyperglycemia, as well as the processes of glucose regeneration from non-carbohydrate precursors (gluconeogenesis) and glycogenolysis under glucose deficiency in the blood. Violations of the liver's glucostatic function can result in T2D: specifically, a decrease in the function of pancreatic beta-cell glucokinase leads to the development of maturity-onset diabetes of the young, MODY2 [12]. There is no data in the literature on the effect of the rs10841855 variant in the glycogen synthase 2 gene on predisposition to T2D in residents of the Slavic ethnic group of Central Russia, necessitating this validation study.
The aim of the study. To analyze the association of the polymorphic variant rs10841855 (G>T) of the GYS2 gene with the risk of T2D in residents of Central Russia.
Materials and methods. The KSMU Regional Ethics Committee approved the study protocol. The study included 1387 T2D patients (527 males and 860 females) with a mean age of 61,2±6,9 years who received treatment at the Endocrinology Department of the City Clinical Emergency Hospital, Kursk, from November 2016 to October 2019. Endocrinologists confirmed the diagnosis of T2D using World Health Organization criteria [9, 10]. As previously described [13, 14, 15], 1281 healthy donors from the Regional Blood Transfusion Station (492 males and 789 females) with an average age of 60.7±5.6 years were invited to participate in the control group. On an empty stomach, all study participants completed a questionnaire to identify the main risk factors and gave 10 ml of venous blood. The phenol-chloroform method was used to extract genomic DNA from whole blood. The GYS2 gene was genotyped for rs10841855 (G>T) using iPLEX technology on a MassArray Analyzer 4 genomic mass spectrometer (Agena Bioscience). The concentrations of carbohydrate and lipid metabolism parameters (glucose, glycated hemoglobin, triglycerides, total cholesterol, low- and high-density lipoproteins) were measured using a semi-automatic biochemical analyzer Clima MC-15 (RAL). The bioinformatics analysis was carried out using the online resources STRING (https://string-db.org) for visual assessment of the GYS2 interactome at the level of protein gene products; GTEx Portal (https://gtexportal.org) to study tissue-specific expression of the GYS2 gene in carriers of variant rs10841855 alleles; and Haploreg v4.1 (https://pubs.broadinstitute.org/mammals/) to study epigenetic regulation of the GYS2 gene. The atSNP resource (http://atsnp.biostat.wisc.edu) was used to investigate transcription factor affinity in the SNP region. The collected data were statistically analyzed with the SNPStats program (https://www.snpstats.net/start.htm). Co-dominant, dominant, recessive, over-dominant, and log-additive genetic models were all tested. As the best model, the one with the lowest numerical value of the Akaike criterion was chosen. At P<0.05, the association was deemed significant. The Kolmogorov-Smirnov test was used to determine the normality of the distribution of biochemical parameters. Non-normally distributed indicators were labeled as median (Median), first (Q1), and third (Q3) quartiles: Median [Q1; Q3]. In such cases, the Kruskal-Wallis test was used to determine statistical significance. The observed differences between groups were considered statistically significant at P<0,05.
Results and Discussion. The studied SNP conformed to the Hardy-Weinberg equilibrium (P>0.05). According to data from the 1000 Genomes project deposited with Ensembl (https://www.ensembl.org), the frequency of the minor allele T of the studied SNP of the GYS2 gene in the Central Russian population was significantly lower than in the European population. Figure 1 shows a diagram of the rs10841855 genotype frequencies.
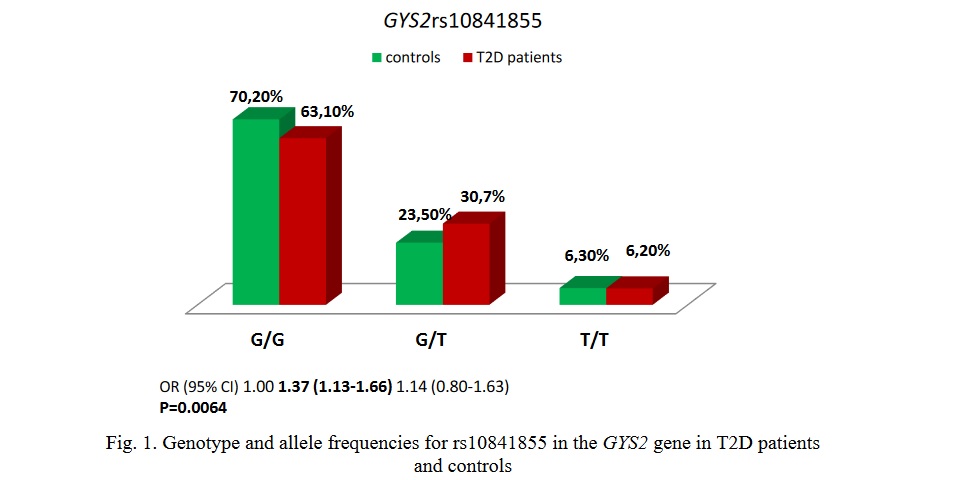
The genotype rs10841855-G/T GYS2 was associated with an increased risk of developing T2D (OR 1.37; 95% CI 1.13-1.66; P=0.0064). The minor allele rs10841855-T (OR 1.25; 95 % CI 1.09-1.43; P=0.001) was significantly more common in the DM2 group (21.6%) than in the control group (18.1%).
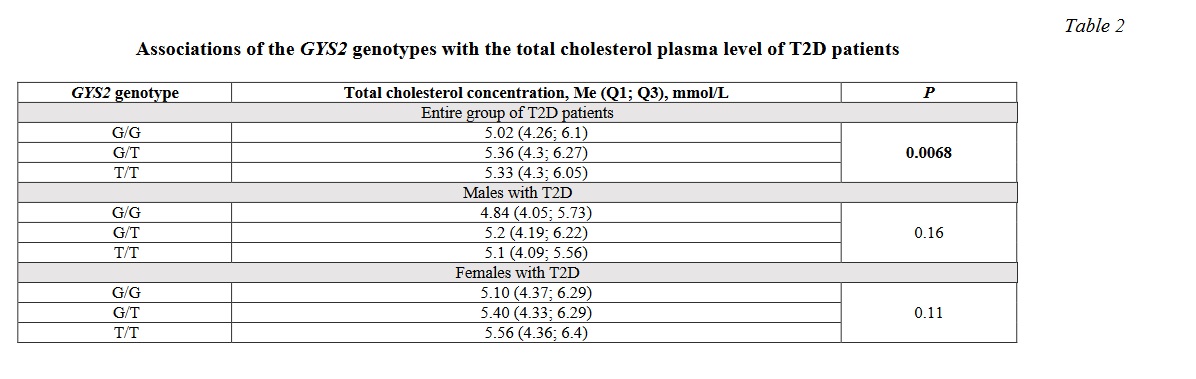
Table 1 shows the results of the sex- and BMI-stratified analysis of the associations of the studied GYS2 polymorphism with the risk of T2D. The association of the genotype rs10841855-G/T (OR 1.71; 95% CI 1.32-2.22; P=0.0001) was found to be typical only for overweight and obese females. The described risk association was not observed in females with normal body weight or in males, regardless of BMI (P>0.05). Furthermore, when the relationship between GYS2 genotypes and biochemical parameters of T2D patients' plasma was examined, it was discovered that carrying the rs10841855-T allele was associated with a higher level of total cholesterol in the blood plasma of T2D patients (P=0.0068, Table 2).
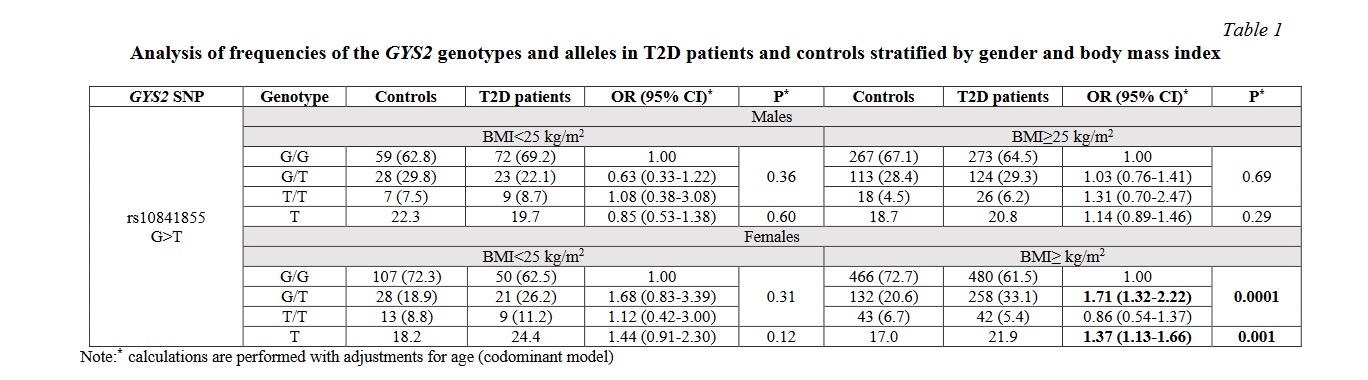
The enzyme glycogen synthase 2 (2.4.1.11) was identified on the metabolic map of carbohydrate metabolism using the KEGG Pathways resource (Fig. 2). The enzyme controls the glycogenesis process, which begins in the liver with the phosphorylation of glucose by glucokinase (2.7.1.2). Phosphoglucomutase converts the reaction product, glucose-6-phosphate, to glucose-1-phosphate (5.4.2.5). UDP-glucosyl pyrophosphorylase (2.4.1.29) uses the energy of one UTP molecule to convert glucose-1-phosphate to UDP-glucose. The enzyme glycogen synthase 2 (2.4.1.11) was identified on the metabolic map of carbohydrate metabolism using the KEGG Pathways resource (Fig. 2). The enzyme controls the glycogenesis process, which begins in the liver with the phosphorylation of glucose by glucokinase (2.7.1.2). Phosphoglucomutase converts the reaction product, glucose-6-phosphate, to glucose-1-phosphate (5.4.2.5). UDP-glucosyl pyrophosphorylase (2.4.1.29) uses the energy of one UTP molecule to convert glucose-1-phosphate to UDP-glucose.
Glycogen synthase is involved in the process at 10-20 glucose residues in the primer chain, catalyzing chain elongation by forming alpha1→4-glycosidic bonds. A branching enzyme (amylo-(1→4)-(1→6)-transglycosylase) that forms alpha1→6 glycosidic bonds creates branch points in glycogen. Glycogen synthase activity is regulated both allosterically and covalently [17]. Simultaneously, insulin is the primary activator of glycogenesis, promoting the dissociation of hepatic glycogen synthase from the intranuclear complex with the regulatory protein and its release into the cytosol for subsequent glucose phosphorylation [18]. Glucose-6-phosphate, a reaction product, is an important allosteric activator of GYS2 [19]. Protein kinase A (PKA), protein kinase C (PKC), AMP kinase (AMPK), glycogen synthase kinase 3 (GSK3), and other kinases phosphorylate the GYS2 enzyme at seven amino acid residues. Insulin inhibits GSK3, which keeps glycogen synthase 2 active (dephosphorylated) [20]. Furthermore, insulin signaling activates PDE3B via PKB, preventing cAMP-mediated phosphorylation of GYS2 [18].
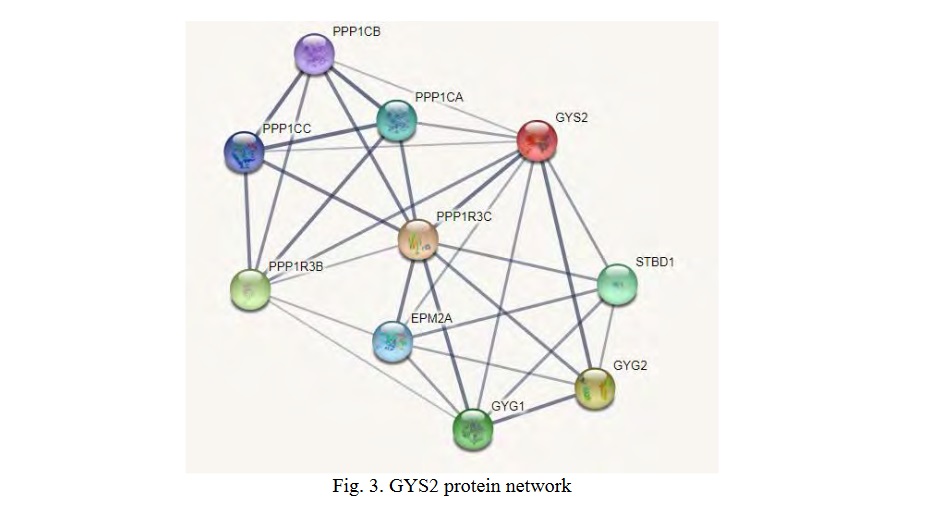
Functional partners of GYS2 (STRING data) form an interactome network of 10 proteins (Fig. 3), including glycogenin 1 (GYG1), glycogenin 2 (GYG2), glycogen-binding domain-containing protein 1 (STBD1), protein phosphatase 1 regulatory subunit 3C (PPP1R3C), regulatory subunit 3B protein phosphatase 1 (PPP1R3B), serine/threonine protein phosphatase 1 catalytic alpha subunit (PPP1CA), serine/threonine protein phosphatase 1 catalytic beta subunit (PPP1CB), serine/threonine protein phosphatase 1 catalytic gamma subunit (PPP1CC), and laforine (EPM2A). According to the Gene Ontology analysis, all of the listed proteins are involved in glycogen metabolism (FDR=3.55×10-22). GYS2, GYG1, GYG2, EPM2A, PPP1R3C, and STBD1 are six network enzymes involved in glycogen biosynthesis (FDR=5.96×10-10), PPP1CA, PPP1CB, and PPP1CC are three proteins involved in the circadian regulation of gene expression (FDR=1.70×10-3).
The current study successfully validated the association between the polymorphic locus rs10841855 in the regulatory region of the GYS2 gene and an increased risk of T2D in residents of Central Russia. The link between this locus and the disease was discovered in a genome-wide study on a European sample of 159208 people [9], where the rs10841855-G allele was linked to an increased risk of developing the disease. The alternative allele rs10841855-T was found to be risky in our study. The rs10841855-T allele has also been linked to a predisposition to T2D in the Japanese population [21]. The literature describes associations of GYS2 gene loss-of-function mutations with the development of glycogenosis type 0 [10] and type 0a [11], in which glycogen synthesis in the liver is disrupted, resulting in ketosis, fasting hypoglycemia with a decrease in lactate and alanine levels, and long-term persistent postprandial hyperglycemia. Increased glycogenesis in the liver of Ankita mice with the T2D phenotype, on the other hand, resulted in decreased hyperglycemia and normalization of liver energy homeostasis [22]. J. Chen et al. discovered a link between GYS2 gene polymorphism and blood plasma insulin levels [23], and a study in the Korean population established an association between GYS2 and obesity, polycystic ovary syndrome, and gestational diabetes mellitus [24].
The current study demonstrated for the first time a link between carrying the rs10841855-T allele and a higher total cholesterol content in the blood plasma of T2D patients, which is explained by the outflow of glucose-6-phosphate accumulating in the liver into lipogenesis, which includes the synthesis of triacylglycerols and cholesterol synthesis.
The rs10841855-T allele is not associated with changes in gene expression in the liver, according to GTEx Portal experimental data. According to the Roadmap Epigenomic Consortium, the rs10841855-T allele is associated with four epigenetic marks in the liver: H3K4me1, H3K27ac in the enhancer region, and H3K4me3, H3K9ac in the GYS2 gene promoter, indicating that the studied variant has significant epigenetic regulatory potential. Interestingly, A. Sharifi-Zarchi discovered that weakly methylated DNA regions are high in H3K4me3, whereas highly methylated genome regions correspond to the H3K4me1 mark [25].
An analysis of transcription factor (TF) binding ability using the bioinformatics tool atSNP revealed that the minor allele rs10841855-T significantly reduces the affinity of three TFs: FOXO1 (P=3,76×10-3), FOXJ1 (P=4,60×10-3), and SOX10 (P=7,29×10-3) preventing them from binding to DNA in the region of that variant allele. It should be noted that FOXO1 in the liver acts as a gluconeogenesis activator [26] and is the primary target of insulin signaling [27].
Conclusion. Thus, the study discovered an association of the GYS2 gene rs10841855 with hypercholesterolemia and an increased risk of T2D in overweight and obese females, which may be due to epigenetic mechanisms of regulation of glycogen synthase 2 gene activity, as well as changes in the network of transcription factors that modulate glucose metabolism in carriers of the alternative allele rs10841855-T. More research, including pharmacogenetic approaches [28], will be needed to determine whether this polymorphism affects the efficacy of type 2 diabetes mellitus treatment.
Financial support
The study was supported by the Russian Science Foundation (N20-15-00227).




















Список литературы