Development of Novel Antimicrobial Peptides Targeting Antibiotic-Resistant Pseudomonas aeruginosa
Abstract
Background: Antibiotic resistance is a global health threat, especially with pathogens like Pseudomonas aeruginosa, which displays high resistance to many antibiotics. Innovative solutions, such as antimicrobial peptides (AMPs), are being explored to combat these multidrug-resistant organisms. The aim of the study:This study aimed to design and synthesize an antimicrobial peptide (AMP) effective against both antibiotic-resistant and susceptible strains of P. aeruginosa, while ensuring safety for human cells. Materials and methods: A novel antimicrobial peptide, WW-185, was designed based on the structure-function relationship of existing AMPs. Its antimicrobial activity was evaluated through Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) assays against standard and multidrug-resistant P. aeruginosa strains. Hemolytic activity was tested to assess safety for human erythrocytes, and time-kill assays were performed to study the peptide’s bactericidal action. Results: WW-185 demonstrated potent antimicrobial activity, with MIC and MBC values comparable to conventional antibiotics. It showed low hemolytic effects on human erythrocytes and exhibited rapid bactericidal action, killing bacteria within 15 minutes and maintaining efficacy for up to 24 hours. The peptide’s activity was attributed to bacterial membrane disruption, facilitated by its structural features. Conclusion: WW-185 is a promising antimicrobial candidate, with potent efficacy against multidrug-resistant P. aeruginosa and low toxicity to human cells. Its potential to reduce rapid resistance development makes it a valuable option for treating antibiotic-resistant infections
Keywords: bactericidal activity, minimum inhibitory concentration, minimum bactericidal concentration, membrane permeabilization, hemolytic activity, WW-185 peptide
Introduction. Pseudomonas aeruginosa, a widespread Gram-negative bacterium, has become a significant opportunistic pathogen, greatly affecting human health [1]. Known for its adaptability and robustness, P. aeruginosa is associated with a wide range of infections, from minor skin and soft tissue infections to serious systemic conditions [2]. This bacterium is especially problematic in causing infections in people with weakened immune systems, such as those with cystic fibrosis, burn injuries, or those undergoing invasive medical treatments [3]. Its capability to form strong biofilms and develop resistance mechanisms makes P. aeruginosa a major clinical challenge, often resulting in hard-to-treat persistent infections [4]. The growing issue of antibiotic resistance in P. aeruginosa strains further complicates treatment, highlighting the critical need for new methods to fight this resilient pathogen [5]. The rising resistance of P. aeruginosa to standard antibiotics is a significant concern in healthcare settings [6]. The bacterium employs various resistance strategies, including efflux pumps, impermeable membranes, and beta-lactamase production, endowing it with an exceptional ability to resist many antimicrobial agents. This resistance not only makes treatment difficult but also stresses the importance of finding new ways to tackle P. aeruginosa infections [7]. Amidst growing antibiotic resistance, antimicrobial peptides (AMPs) have emerged as a promising alternative for dealing with bacterial infections [8]. AMPs are naturally occurring molecules with strong antimicrobial properties, crucial to the innate immune system in different organisms [9]. They are effective against a range of bacteria, including Gram-negative pathogens like P. aeruginosa. AMPs typically attack bacterial membranes, disrupting cell integrity, and often bypassing usual resistance mechanisms. This distinct action mode positions AMPs as a solution for multidrug-resistant bacteria challenges [10]. The need to investigate AMPs as a potential treatment against P. aeruginosa is highlighted by their ability to counteract resistance mechanisms. By attacking bacterial membranes and disrupting vital cellular functions, AMPs could be a strategy against strains resistant to conventional antibiotics [11]. Additionally, the variety of AMPs found in nature offers a vast pool for discovering new compounds specifically effective against P. aeruginosa [12].
The aim of the study. To create new antimicrobial peptides designed to specifically target P. aeruginosa, with a particular emphasis on strains that demonstrate resistance to conventional antibiotics. The objective is to evaluate the effectiveness of these peptides against both antibiotic-resistant and susceptible strains of P. aeruginosa. This study not only enhances our comprehension of antimicrobial peptide actions against P. aeruginosa but also offers potential for the development of novel therapeutic approaches to tackle the urgent problem of antibiotic resistance in clinical environments.
Materials and method
Peptide design and synthesis
The peptide, named WW-158, is a tri-antimicrobial peptide consisting of two tryptophan and one ornithine amino acid units. For this study, the peptide was sourced from GL Biochem Ltd., Shanghai, China, and synthesized using the solid-phase technique in a lyophilized form. Its purification was carried out through reverse-phase high-performance liquid chromatography using a C18 Internsil® column (Thermo Fisher, USA) and an ODS-SP column. The elution process involved an acetonitrile/H2O-TFA gradient at a flow rate of 1.0 mL/min. The purity and identity of the synthesized peptide were verified using electrospray ionization-mass spectrometry [13]. The design strategy aimed to endow the peptide with low cationicity by incorporating the two aforementioned positively charged amino acids [14], facilitating its attachment to the negatively charged bacterial cell membranes via electrostatic interaction. Furthermore, to enhance its ability to permeate the target membranes, the peptide was conjugated with ferulic acid, a highly hydrophobic compound. This molecule is presumed to function as a binding agent, fostering hydrophobic interactions between the peptide and the membrane.
Determination of the Minimum Inhibitory Concentrations (MICs) and Minimum Bactericidal Concentrations (MBCs) for WW-158
The process for determining the Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) of WW-158 was carried out following the microbroth dilution method as prescribed by the Clinical and Laboratory Standards Institute (CLSI) guidelines [15], using sterile 96-well polypropylene microtiter plates. Muller-Hinton broth (MHB) was used as the growth medium to cultivate various bacterial strains, which were initially revived from a frozen glycerol state.
The bacterial cells were grown overnight in MHB and then diluted to a density of 10^6 CFU/mL in the same broth. Various concentrations of WW-158 were prepared, and in separate wells of the 96-well microtiter plates, 50 µL of each peptide concentration was mixed with 50 µL of the bacterial suspension. Each plate had six replicas of every peptide concentration spread over six wells. After an 18-hour incubation at 37 °C, bacterial growth was measured by assessing the optical density (OD) at λ = 570 nm using an ELISA plate reader (Biotech USA). The MIC was identified as the lowest peptide concentration that inhibited bacterial growth. Each plate included a positive control (50 µL of bacterial suspension plus 50 µL MHB without antimicrobial agents) and a negative control (100 µL MHB) to verify bacterial growth and medium sterility, respectively. For MBC determination, 10 µL samples from the clear wells showing no growth and the turbid wells from the positive control were streaked onto sterile, labeled nutrient agar plates. After 24 hours of incubation at 37 °C, the MBC was determined as the lowest concentration at which <0.1% of the original inoculum survived, indicating 99.9% bacterial eradication. All tests were performed in triplicate to ensure result accuracy.
Hemolytic activity of WW-185
The evaluation of WW-185's potential to cause membrane damage in normal erythrocytes was carried out using well-established erythrocyte hemolytic tests, as referenced in previous studies [16]. To guarantee the robustness and reliability of the findings, each test was conducted in triplicate. The formula used to analyze hemolysis is:
% Hemolysis =  × 100
× 100
where A: is OD 450 with the peptide solution,
A0: is OD 450 of the blank.
And AX: is OD 450 of control (0.1% Triton X-100).
MTT Cell Proliferation Assay
The MTT Cell Proliferation Assay was employed in this study using the mammalian Vero cell line obtained commercially from ATCC (ATCC CCL81). Yellow tetrazolium (3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide) MTT was utilized, which, when reduced to purple formazan by reductase enzymes inside metabolically active cells, allowed for the generation of insoluble purple crystals. These crystals were subsequently dissolved in dimethyl sulfoxide (DMSO), and their color was measured spectrophotometrically at a wavelength of 550 nm. For the assay, cells were initially seeded at a density of 5 × 10^3 cells per well in a flat-bottomed 96-well plate, followed by an incubation period of 18-24 hours at 37°C with 5% CO2 to facilitate cell attachment. Subsequently, different concentrations of two peptides and their combination were suspended in RPMI as the dissolving media and added to the cells in the plates at concentrations of 2, 4, 6, 8, and 10 mg/mL for the peptides and 200, 400, 600, 800, and 1000 µg/mL, respectively. An untreated medium was used as a control. The plates were then incubated for an additional 24 hours at 37°C with 5% CO2. After the 24-hour incubation, 20 µL of MTT solution (2.5 mg/mL) was added to each well, and the plates were further incubated for 2-5 hours at 37°C with 5% CO2. Following this incubation, the well contents were removed, ensuring complete removal of all solution. Subsequently, each well received 100 µL of DMSO, which was thoroughly mixed by pipetting to dissolve the formazan crystals until a clear purple color was obtained. The plates were then placed on an absorbance microplate reader (BioTek, Winooski, VT, USA), and the absorbance at 550 nm was measured [17].
Time killing curve
For bacteria, this test has been well standardized and described in M26-A document of CLSI . It is performed in broth culture medium using three tubes containing a bacterial suspension of 5×105 CFU/mL. The first and the second tubes contain the molecule or the extract tested usually at final concentrations of 0.25×MIC and 1×MIC, and the third one is considered as the growth control. The incubation is done under suitable conditions for varied time intervals (0, 4, 6, 8, 10, 12 and 24 h). Then, the percentage of dead cells is calculated relative to the growth control by determining the number of living cells (CFU/mL) of each tube using the agar plate count method. Generally, the bactericidal effect is obtained with a lethality percentage of 90% for 6 h, which is equivalent to 99.9% of lethality for 24 h [18].
Results
Peptide Design and synthesis
The peptide design involved the creation of an enhanced tri-antimicrobial peptide (tri-AMPs) composed of two subunits of tryptophan and one subunit of ornithine amino acids. Ornithine was chosen to confer a positive charge to the peptide. Its uniqueness as an unnatural and non-coded amino acid provides excellent stability against proteases. Tryptophan, selected for its hydrophobic properties and membrane interface interaction, demonstrated a strong preference compared to other hydrophobic amino acids. To further enhance the hydrophobic characteristics of the peptide and exploit its potential antimicrobial activity, it was conjugated to para-hydroxy cinnamic acid
(PHCA). This addition aimed to increase the overall hydrophobicity of the peptide. Notably, PHCA itself possesses inherent antimicrobial properties, thereby contributing to the anticipated enhancement of the peptide's activity. The resulting designed peptide structure was illustrated in Figure 1, showcasing the integration of ornithine, tryptophan, and the conjugated PHCA subunits. This novel peptide design aimed to capitalize on the synergistic effects of its components, combining the stability provided by ornithine, the membrane-interacting properties of tryptophan, and the additional antimicrobial activity conferred by PHCA.
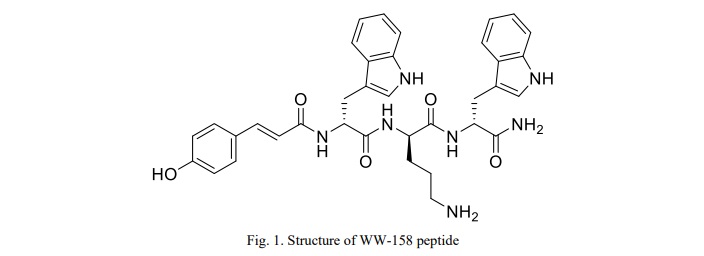
Antimicrobial activity of the peptide
As shown in (Table 1),WW-185 displayed good activity against the standard strain Ps. aeruginosa (ATCC 9027) with an MIC of 6 µg/mL and MIC of 25 µg/mL against MDR Ps. aeruginosa (ATCC BAA-2108). The MBC values were the same as MIC values against the two bacterial types.
Hemolytic activity of the peptides:
The percentage of the red blood cells hemolysis of the peptides alone is shown in Table 2.
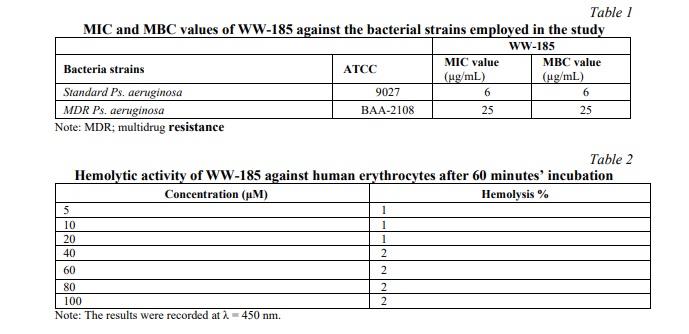
MTT Cell Proliferation
The results of the cytotoxicity assay revealed that the conjugate has an IC50 value of 130 for WW-185 (Fig. 2).
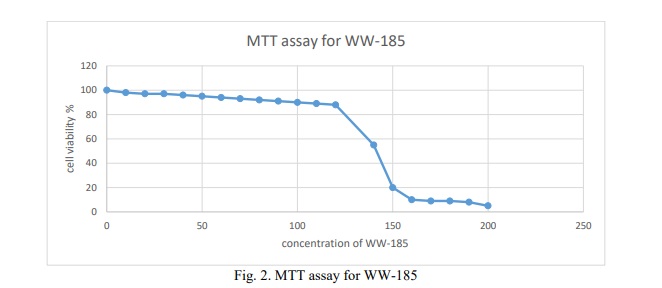
Time kill curve
Results in Figure 3 WW-185 exhibited (no comma) bactericidal effect, reducing the starting log CFU/mL by 3 logs.
Determination of the MIC and MBC of the Individual Antibiotics
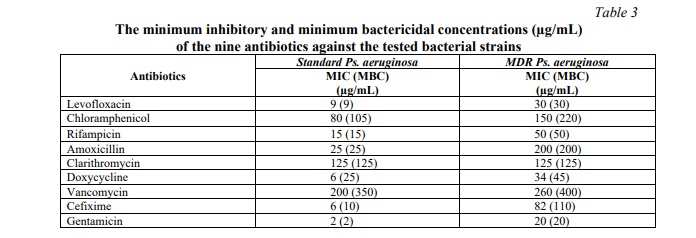
The eight different antibiotics employed in this study were challenged with control and multidrug-resistant strains of Gram-positive and Gram-negative bacteria to determine the antimicrobial activity of each antibiotic against the employed bacterial strains. All the reported data of MIC and MBC values of antibiotics are summarized in Table 3.
Discussion. Antibiotic resistance has evolved into a worldwide health crisis, demanding innovative strategies to counteract multidrug-resistant pathogens [19, 20]. Pseudomonas aeruginosa, a well-known opportunistic pathogen, presents a formidable challenge due to its inherent resistance to numerous antibiotics. In response to this challenge, newly synthesized antimicrobial peptides were developed and specifically tailored to target P. aeruginosa, particularly strains exhibiting resistance to conventional antibiotics. Antimicrobial peptides hold promise due to their broad-spectrum activity and their potential to overcome resistance mechanisms [21].
However, drug development for such peptides has been primarily constrained by safety concerns [22]. This study introduces an approach to peptide design and synthesis, drawing on insights from the structure-function relationship of established AMPs [23]. The effectiveness of these peptides against both antibiotic-resistant and susceptible strains of P. aeruginosa was evaluated in the study. The research revealed that the newly designed peptide, WW-185, exhibits antimicrobial effectiveness similar to conventional antibiotics while having a low hemolytic effect on normal erythrocytes. Both the Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) of WW-185 were on a par with antibiotics from various chemical classes against both standard and multidrug-resistant (MDR) Pseudomonas strains. The identical values of MIC and MBC indicate the bactericidal nature of WW-185. Its microbicidal action primarily stems from the permeabilization of the bacterial cell membrane [24]. The ability of WW-185 to penetrate membranes is likely due to its structure, which features a charge segregation from the hydrophobic center [25]. Consequently, the peptide operates through a unique bacterial membrane disruption mechanism that may be less prone to triggering rapid resistance, suggesting a potentially extended therapeutic application. The study also demonstrated that WW-185, in concentrations up to 100μg/ml, does not cause hemolysis in human erythrocytes, indicating its selectivity for bacterial over host membranes. This finding is consistent with previous research on the peptide 6K-F17, which has shown remarkable bacterial selectivity and non-hemolytic properties towards human erythrocytes at concentrations exceeding 500μg/ml, as reported by Beaudoin et al. (2008) [26]. This specificity may arise because, upon electrostatic attraction to the anionic surface of a bacterial membrane, WW-185's effective hydrophobicity surpasses the threshold for spontaneous membrane penetration. However, in the absence of such attraction, its hydrophobicity remains below the threshold for penetrating mammalian cell membranes, which are zwitterionic. Moreover, the MICs of WW-185 against both standard and MDR P. aeruginosa strains are comparable to those of conventional antibiotics; in some cases, WW-185's MICs are equal to or lower than those of other antibiotics.
The time-kill curve analysis revealed that the bactericidal effect of the peptide was rapid and sustained, exhibiting significant activity against both tested strains in under 15 minutes and maintaining this effect for up to 24 hours. These characteristics are highly sought after in an antimicrobial agent. The peptide's killing action was observed to be time-dependent, aligning with the kinetics of soluble peptides noted in previous research [27]. The mechanism of action for this peptide involves disrupting the cytoplasmic membrane, leading initially to the leakage of small ions and subsequently to the release of larger cellular molecules as the membrane disruption becomes more pronounced [28]. The efficacy of WW-185 against multi-drug resistant (MDR) Pseudomonas aeruginosa is particularly noteworthy. This effectiveness offers fresh perspectives in the battle against MDR strains, underscoring the potential of WW-185 as a valuable tool in antimicrobial therapy.
Conclusion. The creation of novel antimicrobial peptides designed to combat antibiotic-resistant strains of P. aeruginosa represents a crucial advancement in tackling the global challenge of antibiotic resistance. This research not only contributes to the comprehension of antimicrobial peptide mechanisms but also holds significant promise for the development of innovative treatment approaches in the ongoing fight against multidrug-resistant bacteria. The potential efficacy of the synthesized peptides may pave the way for groundbreaking treatment modalities in clinical settings, addressing the critical issue of antibiotic resistance.
Financial support
No financial support has been provided for this work.





















Reference lists