Bioinformatic analysis of microduplications at 5p15.33: identification of TPPP as a candidate gene for autism and intellectual disability
Abstract
Background: Autism is a common psychiatric disorder in children. Since autism is a multifactorial disease, the genetic predisposition plays a significant role in the pathogenesis. However, numerous studies focused on genomic abnormalities in autism are unable to provide reproducible information about pathogenic processes causing this devastating disorder. The aim of the study: The identification of candidate genes by bioinformatic analysis of recurrent copy number variations (CNV) (5p15.33 duplications) revealed by molecular karyotyping in a clinical cohort. Materials and methods: Molecular karyotyping of 296 children with idiopathic autism, intellectual disability was performed by SNP-array. Bioinformatic analysis was made using an original algorithm. Results: Molecular karyotyping genome-wide analysis revealed 3 cases of 5p15.33 duplications. Bioinformatic analysis identified a candidate gene TPPP for brain dysfunction. TPPP is highly expressed in the brain; the gene encodes a protein catalyzing tubulin polymerization, which is important for oligodendrocytes myelination. Interactome analysis was performed to identify pathogenic processes associated with CNV involving TPPP. Expanded TPPP interactome network encompasses 37 proteins, 19 of which are associated with the synaptic plasticity and axonal guidance involved in the normal development and functioning of the brain. Changes in these processes may lead to autism and intellectual disability. Interestingly, clinical genetic databases have not previously associated this gene with a disease condition. Conclusion: Bioinformatic analysis of 5p15.33 CNV allowed us to show that TPPP is a candidate gene for alterations to the development and functioning of the brain. Accordingly, possible disease mechanisms leading to the development of autism with intellectual disability have been proposed. Since data on candidate processes is useful for personalized treatment, we conclude that molecular karyotyping complemented by our original in silico analysis of epigenome, proteome and metabolome is to become an important component for basic and applied research in psychiatric genetics.
Keywords: copy number variants, chromosome 5, bioinformatics, molecular karyotyping, TPPP, autism, intellectual disability
Introduction. Autism spectrum disorders and intellectual disability are common in children and adolescents. Average incidence of autism varies from 1 to 10 cases per 1000 individuals around the world [1, 2]. The main symptoms of autism include speech and communication abnormalities, impaired social adaptation, motor function alterations, and stereotypies. Autism research shows that genetic predisposition plays a significant role in the etiology [3]. Autism may be associated with large chromosomal aberrations (2-10%), copy number variations (CNV) (5-15%) [3-5], and single-gene mutations (5-10%) [4]. In total, genomic pathology affects no less than 25-35% of patients with autistic disorders. However, focusing on autism predisposition genes is limited to expanding the list of candidates. Available data indicate high heterogeneity and low penetrance of mutations associated with this disease. Moreover, there are speculations about reducing autism risks by variations in a number of genes. Apparently, autism is not associated with single genes, but with a variety of molecular and cellular processes [3, 6, 7]. Determining the mechanism underlying the pathogenesis of autism represents an important area of biological psychiatry and medical genetics. Consequently, bioinformatic analysis of genomic variations in individuals with autism and/or intellectual disability to uncover altered molecular and cellular processes is a significant step forward for unraveling mechanisms of the disease and providing the evidence-based therapeutic opportunities.
Aim of the study. Here, we have attempted to characterize microduplications (large CNV) affecting chromosome 5p15.33 by a bioinformatics analysis of molecular karyotyping data.
Materials and methods. Among 296 children with autism, intellectual disability and congenital malformations/developmental delays (age: 2-13 years (average age: 5.5 years); sex ratio: 128/168 (females/males) or 1/1.3), three individuals have been found to demonstrate CNV manifesting as duplications at 5p15.33. Duplications encompassed same genomic loci in three children with autism and intellectual disability. All three children had been diagnosed to have autism, intellectual disability and developmental delays. Molecular karyotyping using Affymetrix CytoScan HD Arrays platform has been performed with an average resolution of ~1000 bp. In silico analysis of the phenotype outcome has been performed using original bioinformatic techniques allowing modulating consequences of genome imbalances at transcriptome, proteome and metabolome levels. The technique has been previously described in details [8-11].
Results and discussion. Clinically, all three children demonstrated an idiopathic autism assessed by the Child Autism Rating Scale (CARS) and intellectual disability. In addition to autistic traits, two children had congenital anomalies. The first patient had a hydrocephalic shape of the skull, lowered outer corners of palpebral fissures. The second had severe microcephaly, a triangular face, wide distal finger phalanges, protruding auricles, a flat bridge of the nose, skin rashes and corpus callosum hypoplasia. The third child demonstrated exclusively wavy hair, which was not observed in other members of his family.
The first patient exhibited 5p15.33 duplication (genomic location: 448542-676847, size: 228305 bp) affecting 7 genes: EXOC3, PP7080, SLC9A3, MIR4456, LOC100996325, CEP72, TPPP. The second patient demonstrated 5p15.33 duplication (genomic location: 448542-819920), size 371378 bp) affecting 8 genes: EXOC3, PP7080, SLC9A3, MIR4456, LOC100996325, CEP72, TPPP, ZDHHC11. The third patient showed 5p15.33 duplication (genomic location: 448542-1175479, size: 726937 bp) affecting 14 genes: EXOC3, PP7080, SLC9A3, MIR4456, LOC100996325, CEP72, TPPP, ZDHHC11, BRD9, TRIP13, MIR4635, NKD2, SLC12A7, LOC100506688. Further characteristics of the duplications are given in Figure 1 and Table 1. No other detectable CNV have been associated with the phenotypic features in these children.
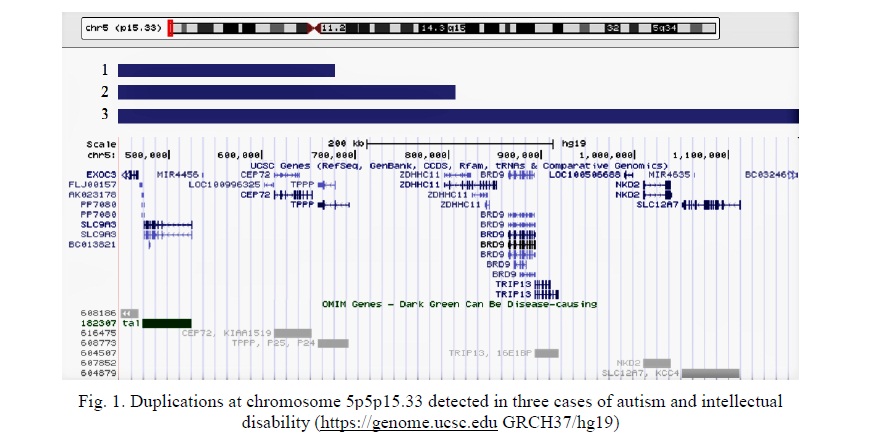
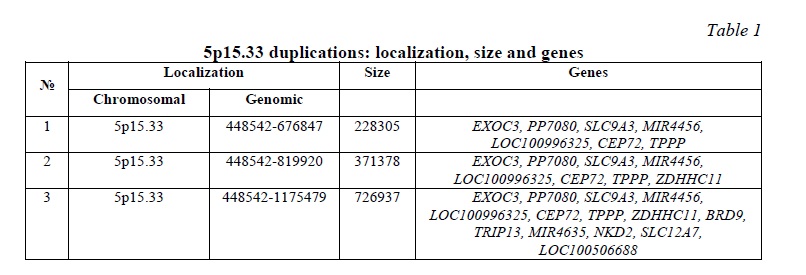
Regardless of differences in length of duplications and the number of affected genes, the overlapping region exists. It is important to note that distal breakpoint is the same in all three cases. Since CNV/duplications result from alterations to recombination, replication, and DNA repair in specific genomic loci [12, 13], one can suggest a recombination hotspot localized at 5p15.33.
Previously, essential genetic processes/changes mediating brain diseases were defined as those occurring directly in cells of the diseased brain [14, 15]. Therefore, in silico gene expression analysis may provide information for prioritizing candidate genes/processes for brain dysfunction [9]. Our analysis has demonstrated that TPPP has the highest expression in the brain as to remaining duplicated genes. Moreover, the relative expression is increased in almost all analyzed areas of the brain (Fig. 2).
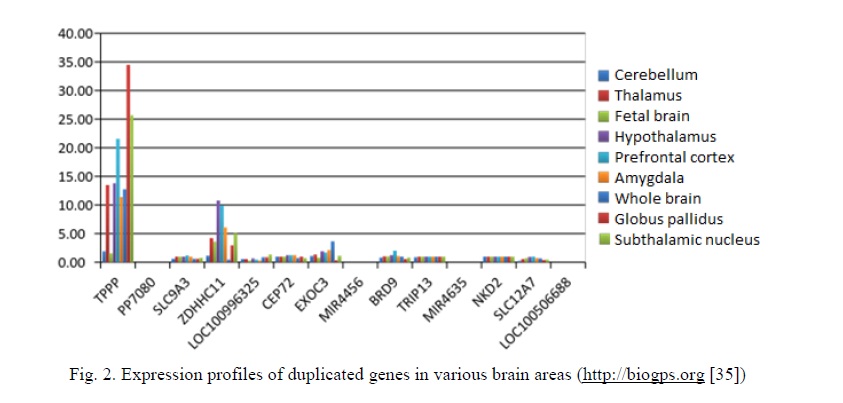
The first duplication affected exons 3 and 4 of TPPP, which encode phosphorylated protein domains, which are involved in the regulation of the protein activity. In the second and third cases, TPPP is completely duplicated. The gene has 4 exons and encodes a protein catalyzing the polymerization of tubulin, a component of microtubules. Microtubules are essential components of cellular processes, such as intracellular transport and cell division (chromosomal disjunction in mitosis and meiosis). In addition, microtubules play a role in myelination of oligodendrocytes [16, 17]. Previously, possible roles of TPPP in the development of neuropsychiatric disorders in children have been suggested [9]. Taking these data into account, TPPP has been associated with autism and intellectual disability.
To gain further insights into mechanisms of phenotypic outcomes of the duplications, an interactome analysis was carried out. As a result, TPPP was found to interact with 7 proteins: SCNA, GAPDH, CDK5, CDK5R1, TMED3, LIMK1, ROCK1 (Fig. 3). These are involved in cell cycle regulation, metabolism, and development of the nervous system. Figure 3 shows TPPP interactome.

To describe ontologies associated with alterations to the interactome due to TPPP duplications, a brief overview of proteins involved in this network is given.
- SNCA (alpha-synuclein) protein belongs to the synuclein family. SNCA expression is high in brain cells. Alpha-synuclein is associated with the membrane of vesicles in neurons. The protein is involved in the control of the transport to presynaptic membrane. SNCA is associated with Parkinson's disease [18].
- GAPDH (glyceraldehyde-3-phosphate dehydrogenase) is an enzyme required for oxidative phosphorylation. It is involved in the nitrosylation of nuclear proteins and regulation of mRNA stability, as well [19].
- Cyclin-dependent kinase 5 (CDK5) — unlike other members of the cyclin-dependent kinase family the protein does not directly regulates of cell cycle. Instead, CDK5 is associated with synaptic plasticity and neuronal migration. This enzyme phosphorylates proteins. The phosphorylation is involved in regulation of cytoskeleton, endocytosis, exocytosis, and apoptosis. CDK5 expression is increased in postmitotic cells of the central nervous system [20].
- CDK5R1 (p35) is a neuron-specific activator of the cyclin-dependent kinase CDK5 by calpain-based proteolytic cleavage of p35 to form the p25 form. CDK5-p25 complexes cause changes in kinase structure and activity [21].
- TMED3 (transmembrane protein p24 containing domain 3) is encoded by TMED3, a gene, which is not indexed in the OMIM database [22].
- LIMK1 regulates actin remodeling by phosphorylating cophilin and converting it into an inactive form. This rearranges dendrite spines and axon modifications to form synaptic plasticity [23].
- ROCK1 (serine/threonine kinase) is a Rho-associated kinase activated by binding between Rho and guanosine triphosphate (GTP). This protein is involved in bioprocesses mediated by modification of the actin cytoskeleton and formation of actomyosin complexes [24].
Apparently, LIMK1 and ROCK1 are involved in pathways of axonal guidance and cytoskeleton regulation. These pathways play a key role in the formation and functioning of the nervous system [25, 26]. The improper regulation causes a decrease in the viability and functional activity of cells, and leads to genomic (chromosomal) instability, which is an element of pathogenic cascades in a wide spectrum of brain diseases [27-29]. Interactome analysis shows that TPPP is involved in brain development and functioning. Therefore, TPPP copy number changes leading to altered gene dosage may have negative effects on neurodevelopmental diseases (i.e. autism and intellectual disability).
Expanded interactome analysis was carried out to highlight ontologies or candidate processes for neurodevelopmental abnormalities associated with 5p15.33 duplications. As a result, 37 proteins were found to interact with TPPP: CDK5R1, GAPDH, SNCA, LIMK1, TMED3, ROCK1, CDK5, RHOA, CCND2, CABLES1, UBC, FYN, ENO1, MAPT, SNCAIP, PARK2, SLC6A3, CDK5R2, NDEL1, PPP1R1B, RAC1, APP, CDC42, MSN, CSNK2A1, RND3, PGK2, HSPA4, RPL13A, PARK7, CCNB1, LRRK2, CDKN1B, DPYSL2, AMPH, MYH14, RHOC (Fig. 4).

According to the largest ontologies, proteins were clustered as follows: proteins associated with the glycolysis enzymes, structural and functional proteins of synaptic connections, proteins involved in actin polymerization and axonal guidance; proteins regulating cell cycle proliferation and cell differentiation (Fig. 4). It should be taken into account that proteins may be simultaneously involved in several processes. Consequently, we have clustered proteins according to ontologies relevant to brain development and functioning. The proteins of synaptic plasticity (I) and axonal guidance (II) are of the interest in the context of brain dysfunction. The first group includes APP, SLC6A3, PPP1R1B, TMED3, SNCA, AMPH, and the second group includes CDKN1B, CDK5R1, CDK5R2, LIMK1, DPYSL2, CDK5, NDEL1, MYH14, RND3, MAPT, CABLES1, MSN, ROCK1. According to the literature, a large number of autism-associated genes encode proteins involved in synaptic plasticity [30]. Axonal guidance is a key mechanism for the development of brain structures during brain development. Genes ontologically associated with axonal guidance are occasionally mutated in autism and intellectual disability [31-34]. It should be noted that 12 out of 37 proteins of the expanded TPPP interactome (GAPDH, HSPA4, PARK7, PARK2, LRRK2, CDK5R2, MAPT, CDK5, APP, SNCA, SLC6A3, PPP1R1B) are elements of the neurodegeneration pathway (Fig. 4, marked with asterisks).
Conclusion. We report on an in silico analysis of functional consequences of 5p15.33 microduplications in 3 children with autism and intellectual disability. Using an original in silico technology modulating phenotypic outcomes of CNV (CNV prioritization) at transcriptome, proteome and metabolome levels, we have found that TPPP is a candidate gene for autism with intellectual disability. Additionally, we have been able to propose a number of candidate processes for neurodevelopmental abnormalities in individuals with 5p15.33 duplications encompassing TPPP. These data are intrinsically useful for forthcoming efforts in developing personalized therapeutic strategies for neurodevelopmental diseases mediated by CNV. Knowledge about consequences of genomic variations generated by identifying candidate processes based on in silico analysis of transcriptome, proteome and metabolome is an important tool for basic and diagnostic genome research.
Financial support
Our study is partially supported by RFBR and CITMA within the research project №18-515-34005. Prof. S.G. Vorsanova’s laboratory is partially supported by the Government Assignment of the Russian Ministry of Health, Assignment no. AAAA-A18–118051590122-7. Prof. I.Y. Iourov’s laboratory is partially supported by the Government Assignment of the Russian Ministry of Science and Higher Education, Assignment no. AAAA-A19–119040490101-6.




















Reference lists