LHCGR gene variants are associated with ovarian response and miscarriage risk following fresh and frozen embryo transfer
Abstract
Background: Currently, infertility is a widespread problem. Despite recent advances in assisted reproductive technologies, success rates still need to be improved. Understanding patients' variability and addressing it with personalized methods may increase the success rates. The aim of the study:The aim of the present study was to analyze the possibility of using variants rs12470652 and rs2293275 of the LHCGRgene to predict ovarian response or pregnancy outcome in assisted reproductive technologies, including the influence of cryopreservation factor. Materials and methods: The present study included 292 infertile women who underwent assisted reproductive technologies. Inclusion criteria were the first or second IVF/ICSI cycle, including women with tubal or/and male factor, anovulation, idiopathic infertility. Exclusion criteria were reduced FSH and/or AMH serum levels. Results: The frequency distribution for rs12470652 and rs2293275 gene variants was AA (94%), AG (6%) and AA (9%), AG (52%), GG (39%) respectively. rs2293275 G allele was found to be associated with low ovarian response. In total, frozen embryo transfer was found to be marginally more efficient, than fresh embryo transfer. In women who became pregnant after fresh embryo transfer, the rs2293275 G allele was statistically significantly found to be a risk factor for miscarriage. Accordingly, frozen embryo transfer hypothesized to be more preferred for rs2293275 G allele carriers to give live birth, than fresh embryo transfer. Meanwhile, fresh embryo transfer might be more effective for rs12470652 G allele carriers to give live birth. Chi-squared test confirmed the results; however, the odds ratio was not applied due to the limited sample size. Further studies and sample increase are required to confirm the results. Conclusion: The present study proposes that LHCGRrs12470652 and rs2293275 genetic variants may improve the prognosis of efficiency of fresh and frozen embryo transfer and might be considered as predictors when planning IVF/ICSI cycles
Keywords: LHCGR, ovarian response, pregnancy, miscarriage, live birth, fresh embryo transfer, frozen embryo transfer
Introduction. Infertility is a global public health issue, affecting millions of individuals world-wide. The etiology of infertility is caused by multiple conditions, including genetic factors [1, 2]. An important representative of the genetic factors class is the gonadotropins receptors genes variation. The luteinizing hormone (LH) and human chorionic gonadotropin (hCG) are two glycoprotein hormones regulating reproductive cycle in women [3, 4]. They share a common α subunit and have a unique β subunit responsible for binding the same LH/hCG receptor (LHCGR) [5]. Despite these similarities, disparate endogenous functions of these hormones are recognized, including quantitatively different early intracellular actions of the cAMP/PKA pathways and steroidogenesis [6]. In fertile age women, LH exerts its best-known functions in the ovary. LH mediates proliferative signals in the granulosa cells, induces internal theca cells to produce high levels of androgens and leads the follicle maturation, the ovulation, the granulosa cells luteinization, the progesterone synthesis, and the corpus luteum maintenance during the luteal phase of the menstrual cycle [1, 7]. hCG is the primate-specific pregnancy hormone produced by trophoblast cells and stimulates the production of progesterone in the corpus luteum, saving the corpus luteum of pregnancy from atresia and mediating the growth of the placenta during pregnancy [1, 6, 7].
LH and hCG bind to a common 675-amino-acid G protein-coupled receptor, characterized by an unusually large extracellular region, which can be subdivided into the high-affinity hormone binding subdomain, and the low-affinity hinge region (LHCGR: L285-E354) involved in the transmembrane-mediated signal transduction [8]. The receptor discriminates between the two ligands and mediates hormone-specific intracellular signaling patterns of cyclic adenosine monophosphate (cAMP)/protein kinase A (PKA)-, protein kinase B (AKT)-, and extracellular signal-regulated kinases ½ (ERK1/2)-pathway activation [6]. LH is more potent than hCG in activating ERK1/2 and AKT phosphorylation, consistently with the requirement of proliferative and survival signals for modulating folliculogenesis. Instead, hCG predominantly triggers steroidogenic events fundamental for maintaining pregnancy and acts mainly through preferential activation of the cAMP/PKA-pathway [7].
The human LH/hCG receptor gene is located on the short arm of chromosome 2 [3] and contains 11 exons [9]. The first 10 exons encode the majority of the extracellular domain, while exon 11 encodes the C-terminal end of the extracellular domain, the entire transmembrane domain, and the intracellular domain [5]. LHCGR exon 10 encodes 27 amino acids within the hinge region (Q290-L316), which is an extracellular segment that is important for hormone selectivity and signal specificity, but not hormone binding [10, 11]. It has been previously described that LHCGR contains evolutionarily conserved residues regulating the hormone–receptor interaction [12]. The murine LH receptor (Lhr) is structurally similar to LHCGR and binds both human LH and hCG. However, Lhr does not discriminate qualitatively between the LH- and hCG-specific signaling [6]. In vitro studies have shown that LHCGR lacking exon 10 can be fully activated by hCG, but with lower LH function than wild-type [10]. The LHCGR exon 10 deletion does not appear to be essential for the hCG or hLH binding, although the native interaction between LH and LHCGR-delExon10 is partially disturbed in a cis-activated process [10, 11], at the same time the absence of exon 10 does not affect either the trans-activation mechanism of hCG or the oligomerization of this receptor variant [11], and also exhibits wild-type-like properties when stimulated with hCG [8]. In addition, previous data indicate that LH may exclusively stimulate the targeted LHCGR through cis-activation, whereas hCG is also able to induce trans-activation [11].
Taken together, it is hypothesized that the presence of LHCGR residues associated with LH and hCG differentiation allows separation of their endocrine signals and optimization of follicular development and pregnancy support. Thus, it is assumed that damage to exon 10 attenuates the LH-mediated ERK1/2 pathway but does not affect the hCG-mediated cAMP/PKA pathway [11]. For these reasons, the two specific LHCGR exon 10 rs12470652 and rs2293275 polymorphisms are good candidate markers for association studies. rs12470652 and rs2293275 genetic variants are located at positions 827 and 935 of exon 10, respectively. In both cases, nucleotide A is replaced by nucleotide G, resulting in the substitution of asparagine for serine (N291S and N312S, respectively) in the LHCGR protein, near glycosylation sites important for signalling regulation [1].
Furthermore, A Delphi Consensus defined that LHCGR polymorphisms may influence ovarian stimulation outcomes and could represent potential future targets for pharmacogenomic research in ART, although data are still very limited [13].
The aim of the study. The aim of the present study was to evaluate the effects of LHCGR variation on ovarian response and assisted reproduction technologies (ART) therapy outcome.
Materials and methods
Study participants. The case-control study included 292 women who underwent controlled ovarian stimulation (COS) for ART treatment at the Center for Human Reproduction and IVF (Rostov-on-Don, Russia) from 2017 to 2019. The women were between 23 and 42 years of age (mean: 32 ± 0.3). All women were undergoing through their first or second ART cycle, including women with tubal or/and male factor, anovulation, idiopathic infertility. Tubal occlusion was diagnosed by laparoscopic examination. Anovulation has been determined by ultrasound as the absence of dominant follicle formation, ovulation, and corpus luteum maturation. Male factor has been identified as the cause of infertility in terms of semen parameters. Due to the predominance of male factor infertility, most of the patients underwent intracytoplasmic sperm injection (ICSI), the rest - by in vitro fertilization (IVF). Exclusion criteria: reduced FSH (<1.8 IU/l) and/or AMH (<0.6 ng/ml) levels. To investigate the association between LHCGR gene variation and ovarian response, all patients were divided into 3 groups according to the number of follicles obtained, which reached a size of 13 mm in diameter in response to COS: normal ovarian response (94 patients with 10-16 follicles); low ovarian response (127 patients with <10 follicles); high ovarian response (65 patients with >16 follicles). To investigate the association between LHCGR gene variation and pregnancy chances, all patients were divided into 2 groups – pregnant after embryo transfer (ET) (n=107) and nonpregnant after ET (n=185). To investigate the association between LHCGR gene variation and miscarriage risk, all patients who had clinical pregnancy were divided into 2 groups – with live birth (n=69) and with miscarriage (n=38). To investigate the impact of the cryopreservation factor, all patients were divided into 2 groups – with fresh ET (n=154) and with frozen ET (n=138).
Informed consent. The present study was conducted in accordance with the principles set out in the Declaration of Helsinki. The protocol was approved by the local Ethnic Committee of the Academy of Biology and Biotechnology of the Southern Federal University (approval certificate № 0104 from 13.02.2017). All women were included after written and verbal informed consent.
Ovarian stimulation and oocyte retrieval. A short protocol of gonadotropin-releasing hormone (GnRH) antagonists was used during COS. Briefly, recombinant FSH treatment was started on day 1 of the menstrual cycle to stimulate follicular growth, GnRH antagonist treatment was started on day 5 for desensitization, ovulation was induced with hCG on day 10. Transvaginal aspiration of follicles under ultrasound control was carried out 36 hours after hCG administration, then the IVF/ICSI procedure was accomplished followed by embryo cultivation and transfer. Due to an enlarged risk of developing ovarian hyperstimulation syndrome (OHSS) in patients with more than 20 follicles obtained, which reached preovulatory maturation in response to COS, embryo transfer was not performed in this cycle, followed by cryopreservation.
Hormone assay. The early-follicular-phase FSH, LH, AMH, progesterone and estradiol levels were determined in the blood serum on the 3rd-5th day of the menstrual cycle, six months before the start of the ART program, using corresponding kits and “Access 2” analyzer (Beckman Coulter, USA).
Genotyping. For DNA isolation and analysis, 10 ml of peripheral blood with EDTA as an anticoagulant was drawn from each patient. Genomic DNA was isolated from blood leukocytes using the DNA-EXPRESS-GENETICS kit (Lytech Co. Ltd., Russia). Genotyping of LHCGR c.935A>G [rs2293275] genetic variant was performed using allele-specific Real-Time PCR with primers: forward primer 5´-GCAACAGCTCCGTAACCAAG-3´, reverse primer 5´-GTGAAAGCACAGTAAGGAAAGTGTA-3´, reverse primer (mut) 5´-TGAAAGCACAGTAAGGAAAGTGCG-3´. Genotyping of LHCGR c.827A>G [rs12470652] genetic variant was performed using allele-specific Real-Time PCR and primers: forward primer 5´-TGCAACAGCTCCGTAACCA-3´, reverse primer 5´-AACCTCTTCTCTTTCAGACAGTA-3´, reverse primer (mut) 5´-AACCTCTTCTCTTTCAGACAGTG-3´. Each reaction was a 25 µl mixture containing 16 µl of deionized nuclease free H2O, 5 µl of qPCRMix (Evrogen Co. Ltd., Russia), 1 µl of each primer, and 2 µl of genomic DNA. PCR reaction conditions were as follows: 5 min denaturation at 94°C, followed by 35 consecutive cycles of 95°C for 15 s, the optimum binding temperature at 59°C for both SNPs, respectively for 10 s, elongation processes at 72°C for 20 s. All samples were validated by double genotyping.
Statistical analysis. Hardy-Weinberg equilibrium (HWE) conformance was calculated applying the Pearson’s chi-square test. Allele frequencies were assessed using the two-tailed Fisher's exact test. The data distribution was detected with the Shapiro-Wilk normality test. Clinical parameters of patients were compared using the Kruskal-Wallis H-test as analysis of variance. A two-sided p value <0.05 was admitted as statistically significant. The associations between genotypes and ovarian response, pregnancy chances and miscarriage risk were computed using the odds ratio (OR) with 95% confidence interval (CI) and chi-square test. Statistical analysis was carried out using the Statistica 13 software.
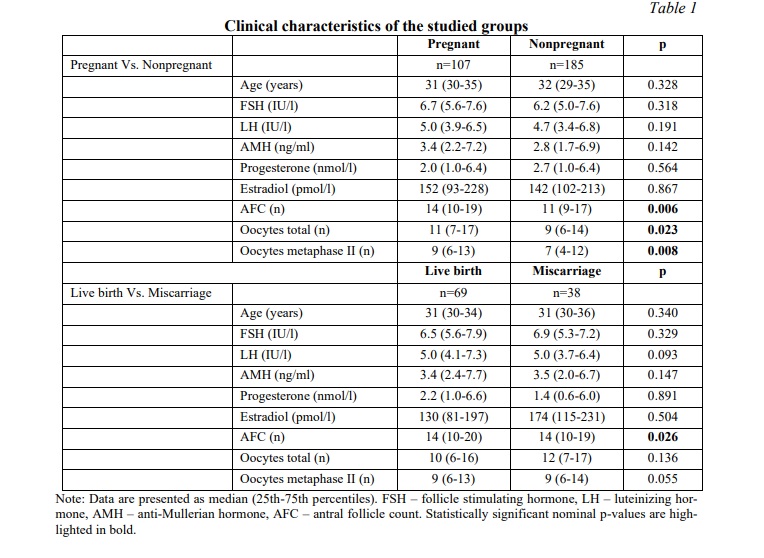
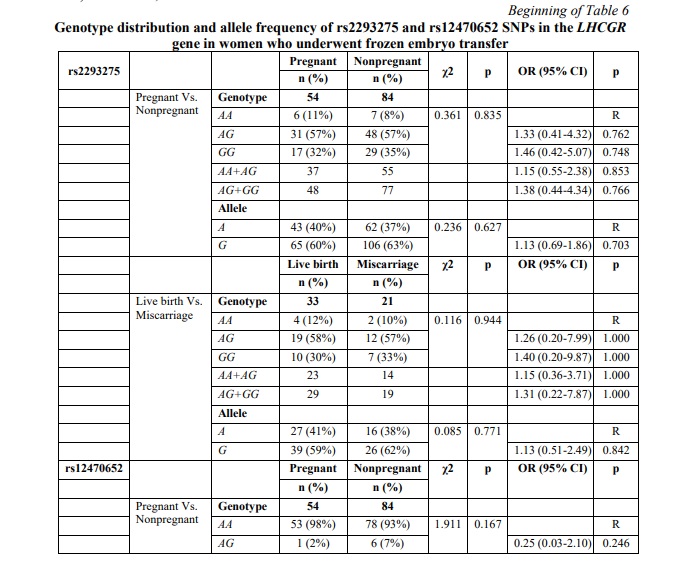
Results. In this study we analyzed the genotype distribution of rs2293275 and rs12470652 LHCGR variants in 292 infertile women undergoing IVF/ICSI procedure aged from 23 to 42 years. Since the human LH/hCG receptor is common for two hormones involved in both folliculogenesis and pregnancy maintenance, the study was carried out in two directions. The first direction was to analyze the association of rs2293275 and rs12470652 LHCGR variants with ovarian response to COS. According to their ovarian response, participants were divided into a “Normal” study group (n=94), a “Low” study group (n=127), and a “High” study group (n=65). The second direction was to analyze the association of rs2293275 and rs12470652 LHCGR variants with miscarriage risk, considering the cryopreservation factor. According to their IVF/ICSI outcome participants were divided into a “Pregnant” study group (n=107) and a “Nonpregnant” study group (n=185). The pregnant study group was further subdivided into two groups (69 women with live birth and 38 women with miscarriage). This study describes cases of both fresh and frozen ET. To assess the contribution of the embryo cryopreservation factor, the studied population was also divided into two groups (154 women with fresh ET and 138 women with frozen ET). Subsequent analyses were performed for the entire studied population, as well as for fresh and frozen ET groups separately.
The SNP rs2293275 in LHCGR gene is associated with ovarian response to controlled ovarian stimulation. The clinical features of infertile women undergoing IVF/ICSI procedure are summarized in Table 1. By conducting the Shapiro-Wilk normality test we determined all data as nonparametric. The median age was 31 years for pregnant women and 32 years for nonpregnant women, and there was no statistically significant difference. Comparing clinical data using the Mann-Whitney U-test, we also found no differences in serum levels of FSH, LH, AMH, progesterone and estradiol either between pregnant and nonpregnant women, or women with live birth and those with miscarriages (Table 1). Nevertheless, ovarian response significantly differed between pregnant and nonpregnant women. AFC (antral follicle count), total and metaphase II oocytes amount was higher in pregnant women (p=0.006, p=0.023 and p=0.008 respectively). However, the subsequent Chi-squared test calculation and odds ratio analysis revealed no statistically significant association either between ovarian response and pregnancy rate or between ovarian response and risk of miscarriage (Table 2). The genotype distribution of the rs2293275 SNP significantly differed between groups of women with normal, low, and high ovarian response (p=0.005). The genotype distribution was inconsistent with HWE in the normal and low response groups (p=0.154 and p=0.986) and deviated from HWE in the high response group (p=0.002). The proportion of wild type homozygous AA, heterozygous AG and mutant homozygous GG in all groups is shown in Table 3. Our results confirmed the association between rs2293275 G allele and low ovarian response (OR (95% CI) = 1.55 (1.05-2.31), p=0.033). The genotype distribution of the rs12470652 SNP was inconsistent with HWE and did not differ between the groups.
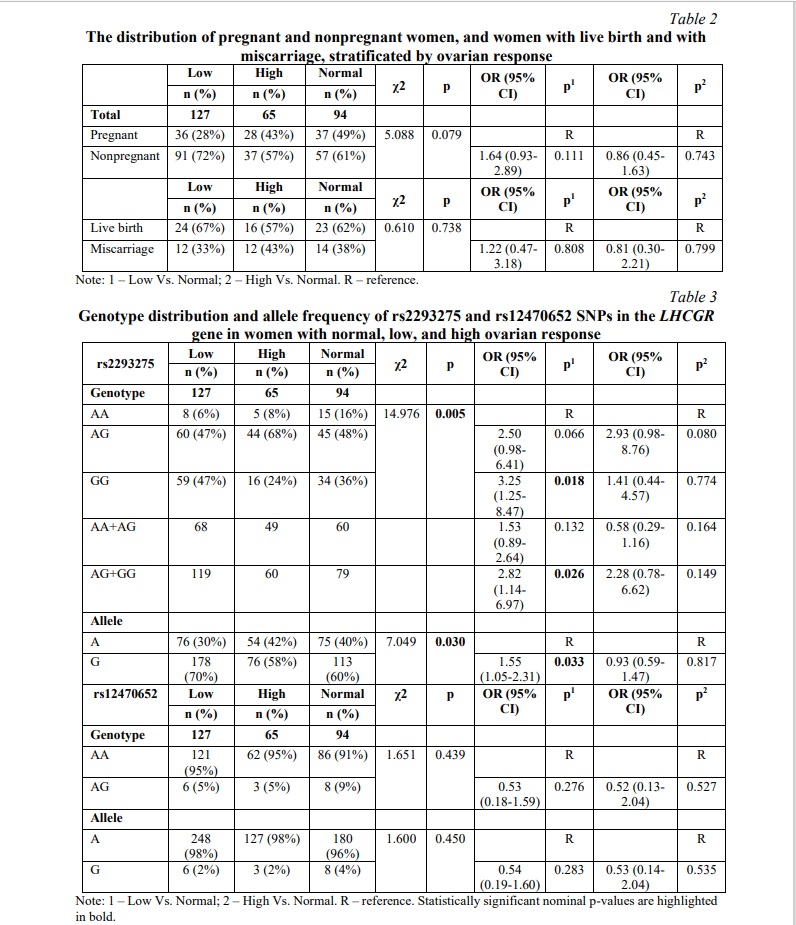
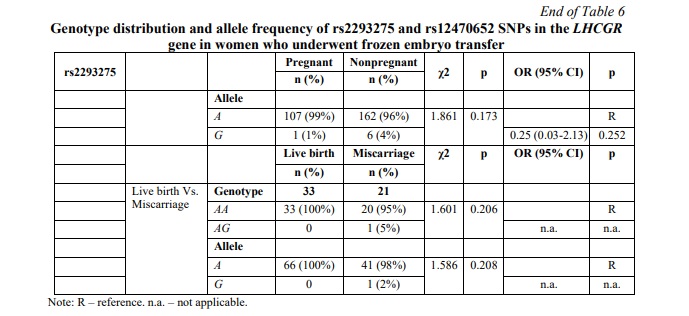
SNP rs2293275 in LHCGR gene is associated with miscarriage risk in women undergoing fresh embryo transfer. The distribution of genotypes for SNP rs2293275 in the pregnant group was inconsistent with HWE (p=0.292) and deviated from HWE in the nonpregnant group (p=0.046). The proportion of wild type homozygous AA, heterozygous AG and mutant homozygous GG was 9, 50 and 41% in the pregnant group, and 10, 53 and 37% in the nonpregnant group, respectively. Our results did not confirm an association between rs2293275 SNP and pregnancy chances for the entire studied population (p=0.810). We also evaluated the association between rs2293275 SNP and pregnancy chances using three genetic models: dominant (AG+GG Vs. AA), recessive (GG Vs. AA+AG), and allelic (A Vs. G). The results revealed no associations (Table 4). To evaluate if there is an association between rs2293275 SNP and miscarriage risk, the pregnant study group was subdivided into a live birth group and a miscarriage group. The genotype distribution of the rs2293275 SNP in both groups was inconsistent with HWE (p=0.357 and p=0.465 respectively). The proportion of wild type homozygous AA, heterozygous AG and mutant homozygous GG was 12, 52 and 36% in the live birth group, and 5, 45 and 50% in the miscarriage group, respectively. Our results did not confirm the association between SNP rs2293275 and miscarriage risk for the entire studied population in any of the genetic models (Table 4). The genotype distribution of the rs12470652 SNP in both pregnant and nonpregnant study groups was inconsistent with HWE (p=0.844 and p=0.620 respectively). The proportion of wild type homozygous AA and heterozygous AG were 96 and 4% in the pregnant group, and 93 and 7% in the nonpregnant group, respectively (Table 4). The mutant homozygous GG were lacking. There was no significant difference in the genotype frequencies distribution between pregnant and nonpregnant women. Our results did not confirm an association between the rs12470652 SNP and pregnancy odds in either the dominant or allelic genetic model. The association between rs12470652 SNP and miscarriage risk was also investigated in the pregnant study group subdivided into the live birth group and the miscarriage group (Table 4). The genotype distribution of the rs12470652 SNP in both groups was inconsistent with HWE (p=0.854 and p=0.934). The proportion of wild type homozygous AA and heterozygous AG were 96 and 4% in the live birth group, and 97 and 3% in the miscarriage group, respectively. There was no significant difference in the genotype frequencies distribution between studied groups. The results did not confirm an association between SNP rs12470652 and miscarriage risk, without adjusting for the cryopreservation factor.
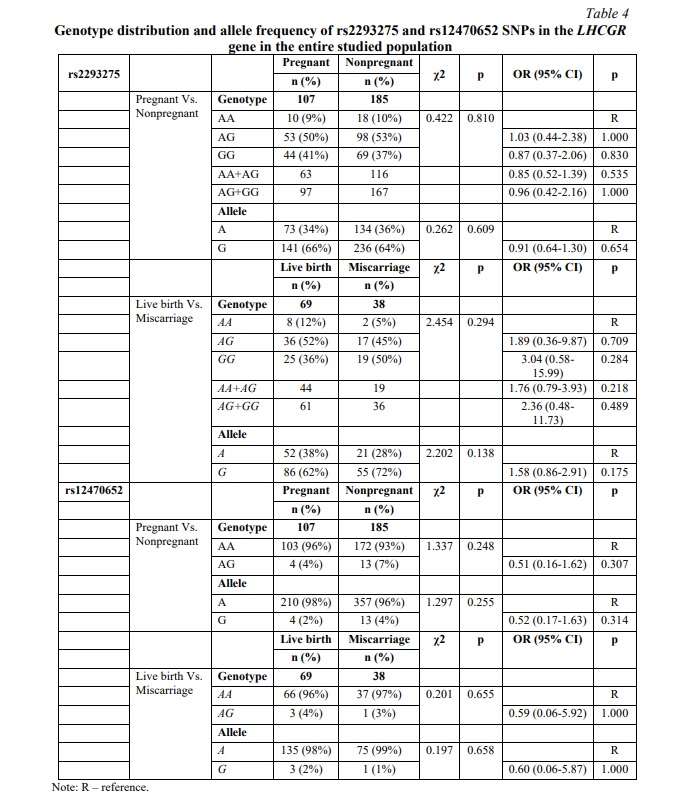
To specifically determine if any associations occur at the expense of the embryo cryopreservation factor, women undergoing fresh or frozen ET were further investigated separately. The results of fresh ET subgroup analysis are clarified in Table 5. No significant differences were observed between the pregnant and nonpregnant cohorts for either rs2293275 SNP or rs12470652 SNP. However, our study revealed an association between rs2293275 SNP and miscarriage in women undergoing fresh ET in the allelic model (p=0.033). According to our results, among women who underwent fresh ET and became pregnant, carriers of the G allele rs2293275 were more than twice as likely to have a miscarriage compared to carriers of the A allele (OR (95% CI) = 3.09 (1.06-8.96), p=0.039).
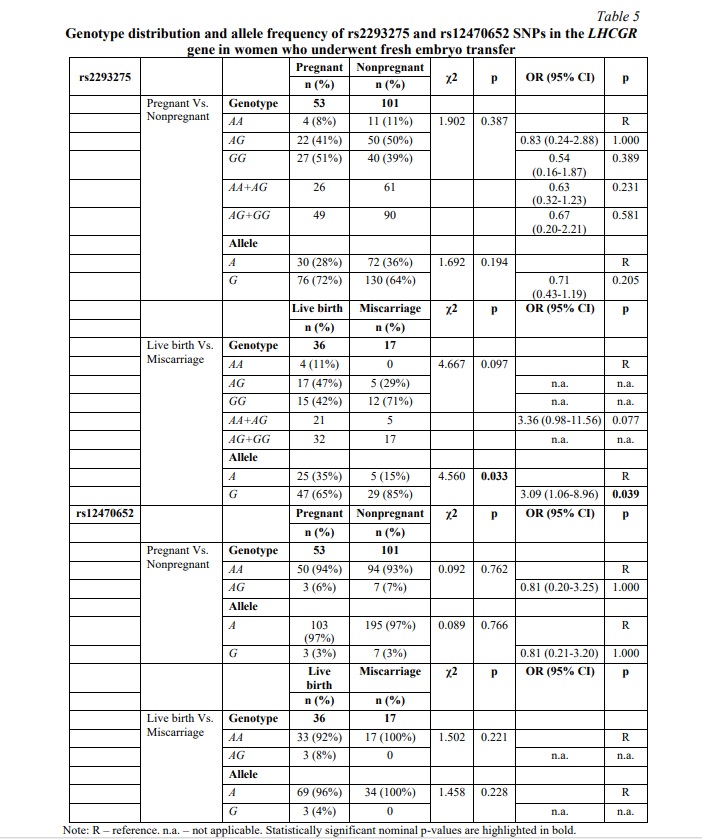
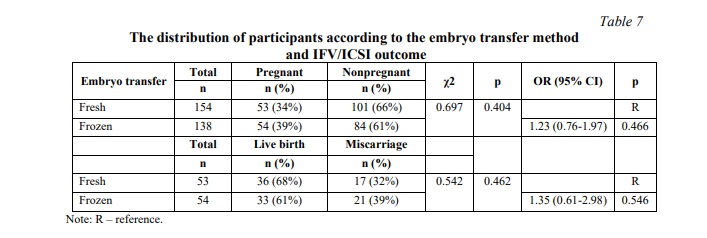
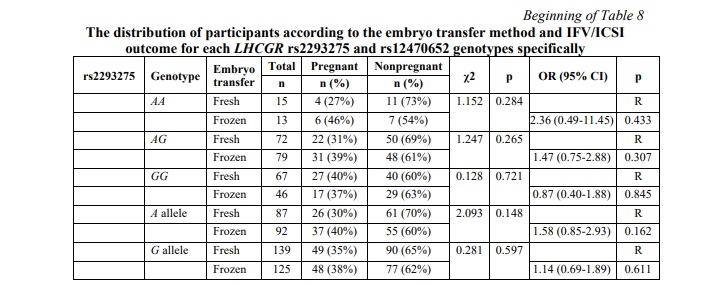
SNP rs12470652 in the LHCGR gene is associated with miscarriage risk in women undergoing a frozen embryo transfer. Further, a similar association research was carried out among women who underwent frozen ET. The results of the frozen ET subgroup analysis are clarified in Table 6. The present study revealed no associations between pregnancy chances and miscarriage risk and any of the studied SNPs in women who underwent frozen ET. To estimate the impact of cryopreservation factor on IFV/ICSI outcome, we compared the distribution of IFV/ICSI outcomes between women undergoing fresh or frozen ET. The results revealed no associations (Table 7). Therefore, the estimation for each LHCGR rs2293275 and rs12470652 genotypes specifically was carried out. Our results revealed an association of the rs12470652 SNP with the risk of miscarriage when comparing women who underwent fresh ET and women who underwent frozen ET, carriers of the AG genotype (Table 8). Among the study participants, who got pregnant, there were only four with the rs12470652 AG genotype. Three of them, who underwent fresh ET, gave birth, and the remaining one, who underwent frozen ET, had a miscarriage. The Chi-squared test confirmed a significant difference (p=0.046); however, the odds ratio was not applied due to the limited sample size.
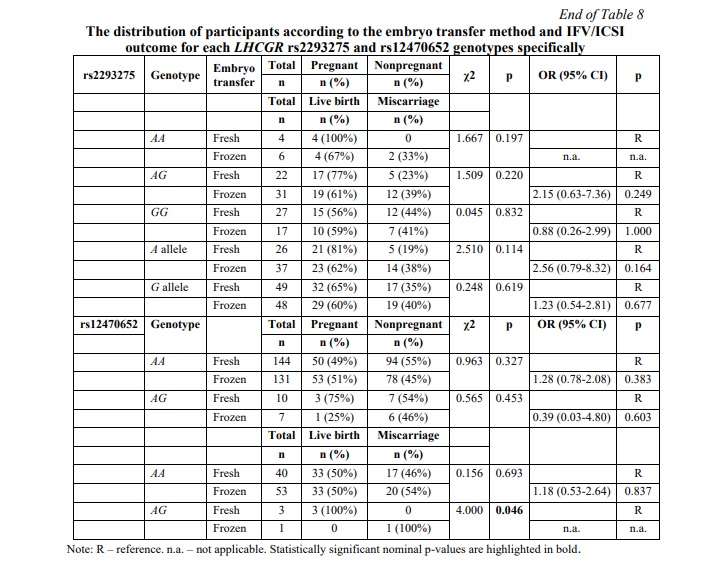
Discussion. LH plays a crucial role in follicle maturation and uterine preparation, and hCG is required to maintain pregnancy. Although they bind the same LH/hCG receptor, their functions, as well as the pathways they initiate, differ [6]. The previously described clinical case report [14] showed that deletion of the LHCGR exon 10, encoding LHCGR hinge region, responsible for hormone selectivity and signal transduction, but not hormone binding [10, 11], does not affect hCG action, while LH action is impaired. The study reported the patient carried an exon 10 deletion affecting the conformational structure of LHCGR, impairing LH- but not hCG-mediated signaling, although maintaining the ability to bind both ligands [14]. This clinical case confirmed the importance of the reported LHCGR hinge region to discriminate between the two hormones. Furthermore, it has been shown that LH activates LHCGR by a cis-mechanism, and hCG-mediated trans-activation is not a common mechanism for both hormones; and the lack of 27 amino acids in the extracellular hinge region of LHCGR encoded by exon 10 affects the cis-activation but not trans-activation mechanism [12].
Furthermore, previous in vitro studies have also shown that cell treatment with LH is accompanied by increased cell viability due to proliferative and anti-apoptotic ERK1/2-pathways activation, while hCG reduced the viable cells number through cAMP/PKA-mediated pro-apoptotic effect [15, 16]. It is also worth considering the diversity of hCG isoforms, including classical hCG, hyperglycosylated hCG, and the free β unit of hyperglycosylated hCG, and sulphated hCG, which have different receptor binding affinities, physiological functions, bioactivity, and effect on implantation [17, 18]. Produced by cytotrophoblast cells during implantation, hyperglycosylated hCG reduces apoptosis of trophoblast cells and induces embryo implantation and trophoblastic invasion [18]. And since rs2293275 and rs12470652 genetic variants are located near the receptor glycosylation sites, this may further interrupt the hyperglycosylated hCG functioning as an invasion promoter and contribute to the pro-apoptotic effect of hCG-mediated cAMP/PKA-pathways signaling. Moreover, hCG could also impact the implantation by modulating the immune balance [17]. Thus, it is hypothesized that the simultaneous disruption due to LHCGR exon 10 variation LH-mediated ERK1/2-pathways signaling and unaffected hCG-mediated cAMP/PKA-pathways signaling, causing abnormalities in circulating levels of several glycoforms of hCG, may lead to growth stagnation and increased miscarriage risk. These assumptions are inconsistent with the results of the present study. Previously C. Guo et al. have shown an association of the rs2293275 GG genotype with higher pregnancy rate after fresh ET [4], and G.A. Ramaraju et al. have also shown an association of the rs2293275 G allele with higher pregnancy rate [19], although it is not clear from this study whether the embryo transfer was fresh or frozen. And vice versa, other studies have shown the rs2293275 AA genotype to be a protective factor for successful IVF, and the GG genotype to be susceptive for IVF failure [20], although it is also not clear from this study whether the embryo transfer was fresh or frozen. Some other studies have shown no associations of LHCGR variants with pregnancy outcome and live birth rates [21]. It is worth noting that the present study showed different associations with miscarriage for both fresh and frozen embryo transfers. No research has previously been conducted to compare the IVF/ICSI outcome between fresh and frozen cycles based on the LHCGR variation. Our findings demonstrated that rs2293275 SNP is associated with miscarriage following fresh ET, while rs12470652 SNP is associated with miscarriage following frozen ET. According to our results, among women who underwent fresh ET and got pregnant, rs2293275 G allele carriers more than twice as often had a miscarriage, compared to A allele carriers (p=0.039). It was also evident that rs12470652 G allele carriers who underwent frozen ET had a higher miscarriage risk when compared to women who underwent fresh ET (p=0.046), although the odds ratio was not applied due to the limited sample size. hCG RNA is already transcribed at the eight-cell stage, the blastocyst produces the protein before its implantation, which subsequently, after embryo transfer, binds to LHCGR on the endometrial surface in the uterine microenvironment [17]. Thus, it is conceivable that embryo freezing alters the blastocyst produced hCG conformation or its RNA transcription conditions, which may interrupt its binding to the receptor in minor rs12470652 allele carriers, causing miscarriage. However, molecular mechanisms remain to be clarified.
The assumption that impaired LH-mediated signaling affects follicles maturation is consistent with our results confirming the association of rs2293275 SNP with ovarian response. The present study revealed the minor rs2293275 G allele to be associated with low ovarian response. Although most previous studies have not found an association of rs2293275 SNP with ovarian response [4, 9, 22], despite a higher prevalence of the minor allele in patients with poor ovarian response [23]. In the present study it was also evident that ovarian response was higher in pregnant women than in nonpregnant. Although it has previously been shown that the delivery rate is closely associated with the number of oocytes retrieved during ovarian stimulation [13], the present study did not confirm the association. This may support the suggestion that hCG has a stronger effect on LHCGR by activation both of cis- and trans-mechanisms, whereas LH activates LHCGR only by the cis-mechanism [12], which is thought to be impaired in G allele carriers.
In total, in the present study the pregnancy rate was 37% for all IVF/ICSI cycles, and the live birth rate was 21%. In fresh cycles the pregnancy rate was 34%, and the live birth rate was 22%, while in frozen cycles the pregnancy rate was 39%, and the live birth rate was 24%. Frozen ET appears to be more efficient than fresh ET, and for rs2293275 G allele carriers even more preferred to give live birth. However, considering the results of the present study, fresh ET might be more effective for rs12470652 G allele carriers to give live birth.
Conclusion. Taken together, both LHCGR rs2293275 and rs12470652 variants were found to be perspective markers in the prediction of assisted reproduction therapy outcomes. Thus, the main findings of the present study were as follows. rs2293275 G allele is associated with low ovarian response. Among women who underwent fresh ET and became pregnant, carriers of the rs2293275 G allele more than twice as often had miscarriage, compared to A allele carriers. There were no significant differences between rs2293275 A and G alleles carriers in outcomes following frozen IVF/ICSI cycles. Accordingly, frozen ET might be more preferred for rs2293275 G allele carriers to give live birth, than fresh ET. Among the study participants, who got pregnant, there were only four with the rs12470652 AG genotype. Three of them, who underwent fresh ET, gave birth, and the remaining one, who underwent frozen ET, had a miscarriage. The Chi-squared test confirmed that fresh ET is more effective for rs12470652 G allele carriers to give live birth; however, the odds ratio was not applied due to the limited sample size. Further research and sample increase are required to confirm the findings.
In summary, according to our data, the prognosis of efficiency of fresh and frozen embryo transfer might be improved if the LHCGR rs2293275 and rs12470652 genotypes are considered as predictors when planning IVF/ICSI cycles.
Financial support
This study was funded by the Ministry of Science and Higher Education of the Russian Federation № FENW-2023-0018 and was performed with the equipment of the Center of collective use «High technologies» (Southern Federal University).





















Reference lists