Association of L432V (rs1056836) polymorphism of the CYP1B1 gene with the increased risk of colorectal cancer in the population of Central Russia
Abstract
Background: Colon malignancies are one of the most common worldwide. Different pro-carcinogenic agents such as polycyclic aromatic hydrocarbons (PAH) and heterocyclic aromatic amines can potentially play a key role in the malignant transformation of cells by interacting with DNA. The enzyme of the 1st phase of xenobiotic biotransformation CYP1B1 is involved in the metabolic activation of various carcinogens. The aim of the study: The aim of our study was to investigate the association between common SNP rs1056836 CYP1B1 and the risk of CRC in the population of Central Russia. Materials and methods: A total of 256 patients with colorectal cancer (134 males, 122 females) and 608 age- and sex-matched healthy controls (279 males, 329 females) were recruited for the study. Genotyping of single nucleotide polymorphism L432V (rs1056836) CYP1B1 were done using Taq-Man-based assays. Results: Single nucleotide polymorphism rs1056836 (substitution L432V) CYP1B1 was associated with the increased risk of colorectal cancer in the population of Central Russia after adjustment for gender, age: ORadj=1.34; 95%CIadj=1.08–1.66; Padj=0.01. Bioinformatic analysis showed the spectrum of transcription factors binding with low-risk C (L) allele are involved in regulation of differentiation of CD8+ alpha-beta T cells (FDR=7.57×10-3) and activation of CD8+ of alpha-beta T cells (P=3.47×10-5, FDR=2.63×10-2). Transcription factors binding with high-risk V allele are not involved in T cell dependent mechanisms of cancer immunoediting. Conclusion: Thus, SNP rs1056836 CYP1B1 is associated with the increased risk of colorectal cancer in the population from Central Russia.
Introduction. Colon malignancies are one of the most common worldwide [1, 2]. Neoplasms with localization in the colon and rectum are combined into the general concept of colorectal cancer (CRC). Colorectal cancer is generally originate from epithelial cells of the intestinal wall and is of multifactorial nature, reflecting the complex interaction of environmental factors with an individual genetic background that significantly causes the risk of the disease [3].
Known environmental risk factors for developing colorectal cancer are the following: high fat diet, alcohol, cigarette smoke, red meat cooked at high temperature; they are sources of high levels of polycyclic aromatic hydrocarbons (PAH) and heterocyclic aromatic amines. These pro-carcinogenic agents can potentially play a key role in the malignant transformation of cells by interacting with DNA [4]. The conversion of carcinogenic compounds entering into the body is carried out thanks to the xenobiotic biotransformation system which is presented in the body with the group of genetically determined enzymes. The enzyme of the 1st phase of xenobiotic biotransformation CYP1B1 is encoded by a gene CYP1B1 and is involved in the metabolic activation of PAH. CYP1B1 plays an important role in carcinogenesis as it activates various carcinogens and it is usually overexpressed in malignant neoplasms. For example, CYP1B1 catalyses the formation of dihydrodiols of specific PAH as well as their subsequent oxidation to carcinogenic epoxides of dihydrodiol [5]. In humans, the CYP1B1 gene includes more than 50 single nucleotide polymorphisms (SNP), one of the most frequent and functionally significant is L432V (rs1056836). Associations between rs1056836 CYP1B1 polymorphism and the risk of colorectal cancer in the population of Central Russia have not been studied yet.
The aim of our study was to investigate the association between common SNP rs1056836 CYP1B1 and the risk of CRC in the population from Central Russia.
Materials and methods. A total of 777 unrelated Russian individuals from the Kursk region was examined; written informed consent was obtained from all participants prior to entering the study. The study included 241 CRC patients (129 males and 112 females) who underwent inpatient treatment at the Kursk Regional Clinical Oncology Center between 2013 and 2017 [6,7]. The control group included 536 healthy volunteers (246 males and 290 females) without a history of chronic diseases. The mean age of CRC patients was 66.89±9.43 years; the mean age of the control individuals was 67.56±6.38 years (P=0.10). The groups (CRC patients and controls) were frequency-matched according to gender (P >0.05).
Patients were included in the group of patients after verification of the final diagnosis of the disease, confirmed by clinical and laboratory-instrumental methods of research (histological examination). The study was approved by the Regional Ethics Committee of Kursk State Medical University. All patients signed a voluntary informed consent to participate in the study. Venous blood samples were obtained from all individuals. Genomic DNA was isolated from peripheral blood by the standard phenol-chloroform extraction method. Genotyping of rs1056836 CYP1B1 polymorphism was performed by real-time PCR using allelic discrimination assay with TaqMan probes on a CFX96 amplifier (Bio-Rad, United States). Regenotyping of 10% of the studied samples taken randomly and in the absence of information on the status of the disease showed 100% reproducibility of the original results.
Associations of alleles with the risk of CRC were performed using logistic regression analysis. To assess association of genotypes with the disease, the odds ratio (OR) and 95% confidence interval (CI) values calculated for the log-additive regression model were used. All calculations were performed, adjusted for gender, age, and smoking in the SNPStats software program, available online (https://www.snpstats.net/start.htm). A value of P≤0.05 was taken as statistically significant.
To study the regulatory potential of the studied SNP, the atSNP online resource was used; it estimates binding affinity of transcription factors (TF) with DNA depending on carriage of reference/alternative allele (http://atsnp.biostat.wisc.edu). TF was included in the analysis only under the condition of a high and very high degree of SNP influence on the interaction of TF with DNA, which was estimated using a position weight matrix. The Gene Ontology tool (http://geneontology.org) was used to search for gene ontologies related to the biological functions of TF associated with the reference/alternative allele.
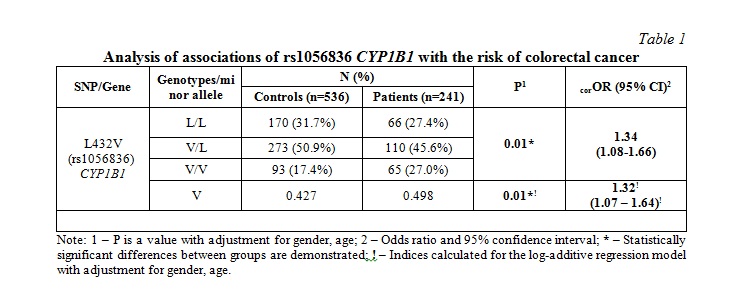
Results and discussion. The genotype frequency distribution of the genotypes rs1056836 CYP1B1 in both control and CRC groups were in Hardy–Weinberg equilibrium (P>0.05). The frequency of the V allele in patients with CRC was higher compared with the control group (OR=1.32; 95%CI=1.07–1.64; P=0.01) (Table 1). A comparative analysis of genotype frequencies also showed that rs1056836 CYP1B1 polymorphism is associated with an increased risk of CRC (Table 1).
The atSNP bioinformatic resource showed that the allele L creates DNA binding sites for 11 transcription factors: ZBTB7B (P=0.001), NFY (P=0.001), ZBTB7C (P=0.003), PAX5 (P=0.005), GLIS2 (P=0.01), RUNX1 (P=0.01), HOXA5 (P=0.02), TBX2 (P=0.03), ELK1 (P=0.04), RUNX1 (P=0.04), AP1 (P=0.05). Allele V creates binding sites for 7 transcription factors: MYC (P=0.003), LBX2 (P=0.004), NFE2L1::MAFG (P=0.02), NFY (P=0.03), EN2 (P=0.04), MYB (P=0.04), Nfe2l2 (P=0.05). Subsequent enrichment analysis of the biological processes using the Gene Ontology Database showed that TF binding with protective C(L) allele are included in the co-regulation of differentiation of CD8+ alpha-beta T cells (P=5.70×10-6, FDR=7.57×10-3) and activation of CD8+ alpha-beta T cells (P=3.47×10-5, FDR=2.63×10-2). Moreover, the carriage of the V allele leads to a loss of DNA binding to TFs involved in the regulation of CD8+ alpha-beta T cells.
It is known that cancer immune-editing (the process due to which the immune system controls tumor growth and forms the tumor immunogenicity) is largely determined by T-cell immunity. Cytotoxic T lymphocytes are one of the most important effector mechanisms of antitumor immunity as they are able to recognize a tumor through their clonal T cell receptors and provide specific destruction of tumor cells by the release of lytic components and direct intercellular interaction [8]. It is remarkable that, cytolytic activity is performed predominantly by CD8+ T-cells [9]. All mentioned above presents important evidence of influence of rs1056836 CYP1B1 on the formation of antitumor immune defenses.
At the same time most of conducted genetic and epidemiological studies have been focused on other association mechanisms of rs1056836 CYP1B1 with colorectal cancer, they reflected functioning and enzymatic activity of the protein encoded by CYP1B1. Taking into account that colorectal cancer is connected with various environmental carcinogens influence and CYP1B1 is a key enzyme of xenobiotics biotransformation and it is highly expressed in the large intestine, we may suppose that rs1056836 CYP1B1 leads to excessive activation of intermediate biotransformation products which are highly DNA damage reagent [10]. CYP1B1 rs1056836 is associated with different substrate specificity and catalytic activity of CYP1B1. It is proved that the variant allele 432V of the CYP1B1 gene determines greater catalytic activity of 4-hydroxylation than a wild-type enzyme [11], it explains the connection of this SNP with increased risk of development of colorectal cancer.
The works devoted to investigation of V432L (rs1056836) CYP1B1 polymorphism involvement for colorectal cancer development are few and contradictory. Thus, the investigations held in the Czech Republic [12] and in Spain [13] did not reveal the associations of rs1056836 CYP1B1 colorectal cancer development. Lack of association of rs1056836 CYP1B1 with colorectal cancer was shown also by two meta-analyzes held in 2012 [14] and in 2014 [15]. At the same time our results are consistent with those of a major study held by The Institute of Cancer Research (Great Britain): they revealed the association of rs1056836 CYP1B1 with colorectal cancer development [16]. We should also note the recent work by Le Morvan et al., which was held on tumor cell lines, and it revealed higher proliferative and invasive activity of tumor cells upon expression of the variant form of rs1056836 CYP1B1, and alsoin vivo. Besides, it was found out that resistance to antitumor drugs significantly increases under the V allele inheritance [17].
Conclusion. Thus, our investigation has revealed an association of the rs1056836 CYP1B1 single nucleotide variant with increased risk for colorectal cancer in the population in Central Russia. The mechanisms of this association may be related to the influence of rs1056836 on functional effects of CYP1B1 which reflect its participation in the activation of xenobiotic with procarcinogenic effects as well as to the involvement of this SNP in the mechanisms of regulation of antitumor T-cell immunity.




















Reference lists
Petrash EA. Social identity disorders in patients with colorectal cancer (colostomy). Kursk Scientific and Practical Bulletin "Man and His Health". 2015;(2):120-125. Russian.
Jeon J, Du M, Schoen RE, et al. Determining risk of colorectal cancer and starting age of screening based on lifestyle, environmental, and genetic factors. Gastroenterology. 2018;154(8):2152‐2164. DOI: http://dx.doi.org/10.1053/j.gastro.2018.02.021