First molecular cytogenetic characterization of the MMT 060562 murine breast cancer cell line
Abstract
Background: Murine cell lines are working horses applied as model systems in multiple research studies in many laboratories. Nonetheless, most of them are not well characterized at the genetic level. This diagnosis holds also true for the MMT 060562 murine breast cancer cell line, also referred to as MMT060562 or MMT-060562. The aim of the study: To provide detailed cytogenetic characterization of the MMT 060562 cancer cell line. Materials and methods: The cell line was studied by molecular cytogenetics, namely by fluorescence in situ hybridization applying all murine while chromosome paints in one probe set and all chromosome-specific murine multicolor banding probe sets. Results: For this we present here the first detailed karyotype of the established in 1962 cell line, a comprehensive map of chromosomal imbalances and an in-silico translation of the results to the human genome. Surprisingly, MMT 060562 has only few chromosomal aberrations, even a cell clone without any gross chromosomal abnormalities in ~40% of the cells, and with most aberrant of four cell clones showing a dicentric dic(2;17)(A1A1), a derivative del(3)(A3C), a trisomy 6, and a derivative der(14)t(13;14)(14pter→14D1::14B→14D1::13A3→13qter). Conclusion: It could be shown, that MMT 060562 is most similar to human breast cancer of basal-like tumor type. Thus, this cell line can serve as a model for a very early breast cancer stage and thus closes a gap in the yet available cell lines.
Introduction. In female breast cancer (BC), being a major leading cause of human cancer death, survival chance varies still tremendously, depending on many factors like awareness, preventive programs, diagnostic regimens, and possibilities to treat this disease [1]. The incidence of this malignancy is furthermore influenced by such factors as average life expectancy in the country the person is derived from, life style, individual estrogen-levels, number of children of the woman, and family history of cancer, including adverse gene mutations [1, 2]. As BC is heterogenic, it is divided into subtypes according to morphology, molecular profiles, and specific biomarker expression [3]. Accordingly, various subtypes may have different prognoses, clinical outcomes, and maybe in need of specific treatment regimens [3, 4]. Biomarkers studied to perform BC subclassifications may include oncogene and tumor suppressor gene expression levels, as well as that of other gene-products, like estrogen-, progesterone-, human epidermal growth factor-2- (HER-2/ ERBB2) and epidermal growth factor-receptor, or cytokeratin 5 or nuclear protein Ki67 [5, 6]. Thus, BC may be classified in (a) luminal A-like, (b) luminal B-like (HER2-positive and HER2-negative), (c) HER2-overexpressing, and (d) triple-negative subtypes [3, 6]. Furthermore, liquid biopsy plays an increasingly important role in BC follow-up [7].
Various treatment regimens for BC subtypes are available [6, 8], however, severe side effects cannot be excluded in many of these approaches. Also, aggressive courses of disease and limitations in BC treatment are not unusual [9]. Therefore, research on BC-biology, as well as for new treatment strategies is imperative [10], which are undertaken in cell cultures based on murine BC cell lines [5].
The MMT 060562 murine BC-cell line, also referred to as MMT060562 or MMT-060562, has been applied in about 2 dozen studies (see [11]), and, as most of other such cell lines [12], still been not characterized in detail genetically. According to ATCC, this cell line was established in 1962 [13] as being derived from a hybrid female C57BL/6xAf mouse as a spontaneous malignant neoplasm of the mouse mammary gland [14]. To the best of our knowledge the karyotype of this cell line was never published before, however, according to ATCC, MMT 060562 has a modal number of 40 chromosomes with a range of 36 to 81, and a stem line number being diploid [13], while ECAAC gives a description that the cell line is diploid with 2n = 40 [14]. To close the gap in the literature, here we present the first molecular cytogenetic characterization of the MMT 060562 cell line.
Material and Methods
Cell line. The MMT 060562 cell line was purchased from the American Type Culture Collection (ATCCR CCL-51™; Wesel Germany) and grown adherently according to the company’s instructions: the cells were cytogenetically worked up as previously described [12].
Molecular cytogenetics. FISH was done as previously reported [15] applying whole chromosome paints (“SkyPaintTM DNA Kit M-10 for Mouse Chromosomes”, Applied Spectral Imaging, Edingen-Neckarhausen, Germany) for multicolor-FISH (mFISH), and murine chromosome-specific multicolor banding (mcb) probe mixes for FISH-banding [16]. At least 30 metaphases were analyzed for each probe set (Zeiss Axioplan microscopy, equipped with ISIS software (MetaSystems, Altlussheim, Germany).
Data analyses. Imbalances and breakpoints of MMT 060562 were determined according to mcb data and aligned to human homologous regions using the Ensembl and the UCSC Genome Browser, as previously described [12]. The obtained data were compared to genetic changes known from human BCs as previously done [5].
Ethics Statement. According to the Ethical Committee (medical faculty) and the Animal Experimentation Commission of the Friedrich Schiller University, there are no ethical agreements necessary for studies involving murine tumor cell lines like MMT 060562.
Results. MMT 060562 is a cell line with nearly stable diploid karyotype, only few single cell aberrations were present, which are not reported here. The aberrations detected could be divided in two clones, with clone 2 having three sub-clones.
- Clone 1 can be considered as the ancestor and has a completely normal murine karyotype representing 42% of the studied metaphases; karyotype: 40,XY.
- Clone 2, falling in three subclones, has as typical aberration in common an additional chromosome 6:
- clone 2a showed a trisomy of chromosome 6 and an inversion in a chromosome 2 (11.3% of the cells); karyotype: 41,XY,inv(2)(C3E5),+6;
- clone 2b1 (28.5% of the cells) acquired a complicated aberration involving chromosomes 13 and 14; 41,XY,+6,der(14)t(13;14)(14pter→14D1::14B→14D1::13A3→13qter);
- clone 2b2 (18.2% of the cells) acquired an additional aberration involving chromosomes 2 and 17 and showed the karyotype 40,XY,dic(2;17)(A1A1),del(3)(A3C),+6,der(14)t(13;14)(14pter→14D1::14B→14D1::13A3→13qter) (Fig. 1).
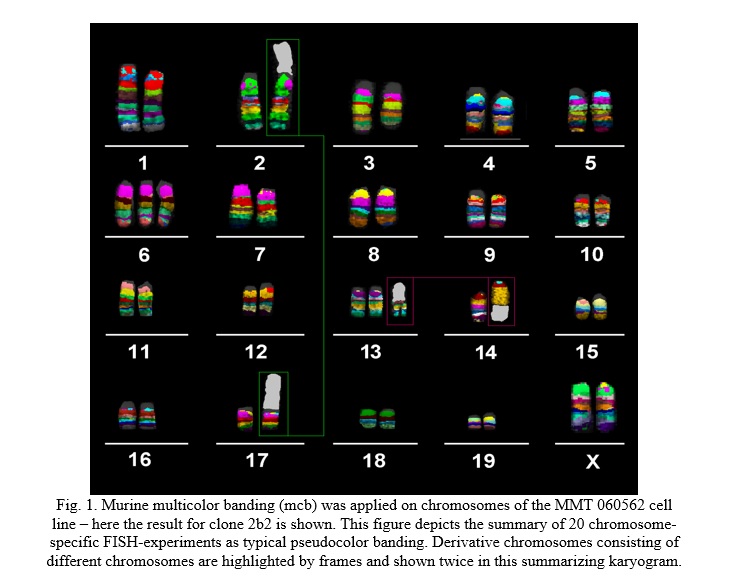
FISH-data translated to CGH-data is summarized in Table 1 and Fig. 2A. An in-silico translation of those results to the human genome identified the corresponding homologous region in the human genome (Table 1, Fig. 2B).


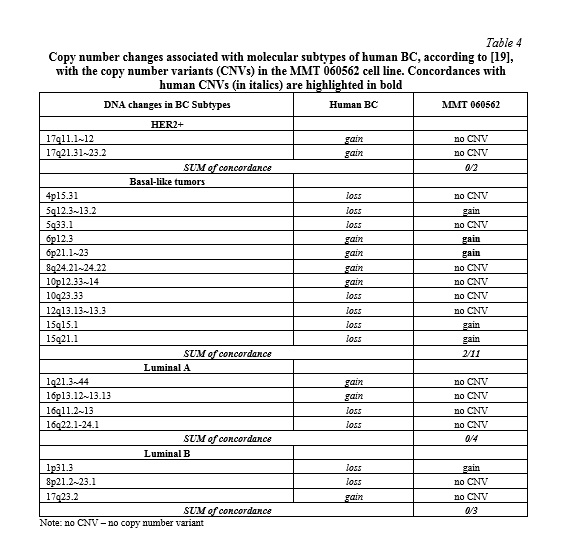
Comparison with literature. The corresponding homologous regions of the MMT 060562 cell line compared with the common imbalances in related to human BC [5, 17, 18] revealed copy number variations in 9 of 21 regions (43%) known to harbor oncogenes and tumor suppressor genes (Table 2). The here reported breakpoints of MMT 060562 compared to chromosomal breaks of human BC presented a congruency of 6%, only (Table 3). The genetic alterations in the cell line correlated with the molecular subtype for human BC as shown in Table 4. The results revealed the best correspondence between MMT 060562 and human BC subtype basal-like tumors (18%).



Discussion. Heterogeneity of BC is one of the reasons that its biology is overall still poorly understood. Correspondingly, basic research and studies testing new potential therapeutics are necessary [9; 20]. MMT 060562 is a murine BC cell line yet not characterized cytogenomically in detail. Thus, its reluctant use in research studies, according to PUBMED [11] only ~20 papers are published using this cell line, here we did the first detailed molecular cytogenetic study to close this gap. The same approach as previously undertaken for several other murine cell lines was done for MMT 060562 [5, 12, 15, 16, 21-23].
The MMT 060562 cell line presents a normal karyotype in ~40% of the studied cells, which is surprising for an almost 60-year-old cell line. Most likely it would be an ideal candidate to be studied by sequencing, to find submicroscopic mutations leading cells on the path towards BC-malignization. Also it is striking that since its establishment MMT 060562 cells did not tertaploidize, as reported mainly for human cell lines [24] and ~50% of murine tumour cell lines [5, 12, 15, 16, 21-23]. To the best of our knowledge, the MMT 060562 cell line is the less chromosomal aberrant malignant cell line ever reported. However, it is definitely a cell line, which induced tumors in nude mice [25].
Conclusion. Overall and in conclusion, the MMT 060562 cell line is a very interesting model system for early human BC, which should be studied in more detail and applied in corresponding studies for new therapeutica.




















Reference lists