Molecular cytogenetic and cytopostgenomic analysis of the human genome
Abstract
Background: Despite the achievements of human genomics, comprehensive genome analysis, including acquiring the knowledge about intercellular and interindividual variations at (sub)chromosomal/cytogenomic level, remains a difficult task. This basically results from a lack of heuristic algorithms for uncovering (cyto)genomic and/or somatic genome variations and the functional outcomes. However, current developments in molecular cytogenetics and “cytopostgenomics” may offer a solution of the problem. The aim of the study: To present a heuristic algorithm for molecular cytogenetic and cytopostgenomic analysis of the human genome to uncover mechanisms of genetic (brain/neurodevelopmental) diseases. Materials and methods: Data on cytogenetic and (cyto)genomic variations (chromosome abnormalities, chromosome/genome instability, copy number variation (CNV) etc.) addressed by original molecular cytogenetic techniques and processed by original bioinformatic (cytopostgenomic) methods were used to develop the algorithm. Karyotyping was performed in 8556 individuals. FISH analysis was applied when required (cases of somatic mosaicism/chromosome instability). Molecular karyotyping by SNP-array was performed in 600 (~7%) cases. Results: Using our long-term experience of studying chromosomal and genomic variations/instability in neurodevelopmental disorders as well as original developments in (cyto)genomic data processing, we managed to present a heuristic algorithm for molecular cytogenetic and cytopostgenomic analysis of the human genome to uncover mechanisms for brain diseases. Estimated efficiency of the algorithm was established to achieve 84%. Analyzing the dynamics of applying cytogenetic and cytogenomic techniques throughout ~35 years of our diagnostic research we found that the diagnostic efficiency had been increasing from ~7% (exclusive diagnosis by karyotyping) to more than 80% (molecular cytogenetic and cytopostgenomic analysis). Conclusion: Here, we propose a heuristic algorithm for molecular cytogenetic and cytopostgenomic analysis of the human genome to uncover mechanisms for genetic diseases. The efficiency and ability to uncover mechanisms of chromosome instability allows us to conclude that the algorithm may be highly competitive for basic and diagnostic genomic/cyto(post)genomic research.
Keywords: chromosome, chromosomal instability, genome, genome instability, molecular cytogenetics, human disease, brain disease, bioinformatics, neurodevelopmental disorders, cytogenomics
Introduction. Molecular cytogenetic and cytogenomic analyses have become an important part of diagnostic and basic research focused on uncovering genetic mechanisms of a disease [1-4]. Brilliant discoveries of novel genetic mechanisms for brain disorders [5, 6, 7] obliged to reevaluate diagnostic workflows and approaches to identification of genomic variations associated with a disease. Nonetheless, there are still numerous problems associated with cytogenomic diagnosis roughly referred to genome mapping, difficulties of interpretation of genome variations (i.e. copy number variation or CNV), and understanding the meaning of somatic genome variations [3, 8, 9]. On the other hand, systems analysis applied to cytogenomic data seems to be able to solve these problems [10]. Additionally, molecular cytogenetic and genetic techniques (e.g. fluorescence in situ hybridization or FISH and genome scanning methods) have a number of limitations, i.e. analysis of specific genome loci (FISH), lack of reproducible interpretation and moderate cell scoring potential (genome scanning methods) [11], which are, however, may be also overcome by the application of bioinformatic or “cytopostgenomic” methods [12]. Furthermore, these methods are required for complementing the knowledge about genome variations [13], i.e. genome behavior at the interindividual [14] and at intercellular levels [15]. Accordingly, it is highly likely that combinations of molecular cytogenetic, cytogenomic and bioinformatic techniques may underlie approaches to uncovering disease mechanisms [16-19].
To succeed in unraveling genomic mechanisms of genetic diseases, genome data are to be processed for estimating functional outcomes of changes in genetic material [16, 20]. More precisely, these data are to be used for uncovering molecular and cellular pathways to a disease [20]. In the molecular cytogenetic/cytogenomic context, these studies have to address somatic chromosomal mosaicism and chromosome/genome instability in addition to disease pathways [15, 20-25]. Furthermore, cytogenomic variations are generally more complex than those affecting single genes [26, 27]. In total, there is a need to address all the variations detectable in an individual genome (variome) with respect to mosaicism and genomic/chromosomal instability for understanding causes and consequences of cytogenomic variations [28]. This becomes even more significant, taking into account the ability of chromosome instability or genome chaos to produce pathological conditions (e.g. cancer and neurodegeneration) through the lifespan [25, 29-32]. Finally, the systems biology methodology (bioinformatics or systems genomics) [33, 34], applied to cytogenomic and cytogenetic data should be efficient for uncovering genetic/genomic mechanisms of a variety of clinical conditions [10, 28, 35] leading to a possibility of treating diseases associated with chromosome imbalances, which are generally considered as incurable [36]. In conclusion, we suggest that combining all the aforementioned methods and may provide a heuristic algorithm for unraveling cytogenomic mechanisms for genetic brain diseases.
The aim of the study. Here, using our previous developments in molecular cytogenetics and cytopostgenonics and analysis of data on karyotyping of 8556 individuals with neurodevelopmental disorders and congenital malformations, we have presented a heuristic algorithm for unraveling (cyto)genomic mechanisms for genetic diseases.
Materials and methods. Karyotyping data, acquired as described previously [37, 38], was obtained during a long-term study (from 1985 to present) of 8556 individuals with neurodevelopmental disorders (intellectual disability, autism, epilepsy, behavioral abnormalities) and congenital malformations. FISH was applied in cases of somatic mosaicism and/or chromosome instability according to previous protocols of hybridization and detection [37-39]. FISH data were acquired from Vorsanova et al. 2021 and 2022 [37, 38]. Molecular karyotyping by SNP array was performed in 600 cases from the karyotyped cohort. The protocol of SNP array was repeatedly described previously [40-43]. Bioinformatic analyses were carried out by an original methodology, which included systems genome (variome) analysis, data fusion, “CNV laundering” and pathway-based prioritization of genomic variations. All these techniques were recently described in detail elsewhere [16, 20, 28, 44, 45].
Results and discussion. Previously, we proposed that karyotyping (cytogenetic analysis) is the initial step of genomic data analysis, which is required for understanding the variability and behavior of an individual genome [17]. More precisely, it was recommended to obtain two data sets: cytogenetic and genomic (cytogenomic). This proposal was lately supported by Breman and Stankiewicz, who noted that karyotyping is to be applied at the start of diagnostic genome research [46]. Figure 1 schematically demonstrates this idea.

Taking into account the efficiency of combining FISH and SNP array (Fig. 2) [36-40], we propose to use these techniques for analyzing cytogenomic variations. The idea is even more apposite for cases of chromosomal instability and mosaicism as well as aneuploidy/polyploidy cases, inasmuch as it gives an opportunity to uncover the molecular and cellular causes of the instability in addition to detection intercellular variations caused by genome chaos (i.e. description of the nature of chromosomal instability) [6, 8, 10, 17, 25, 28, 30, 31].
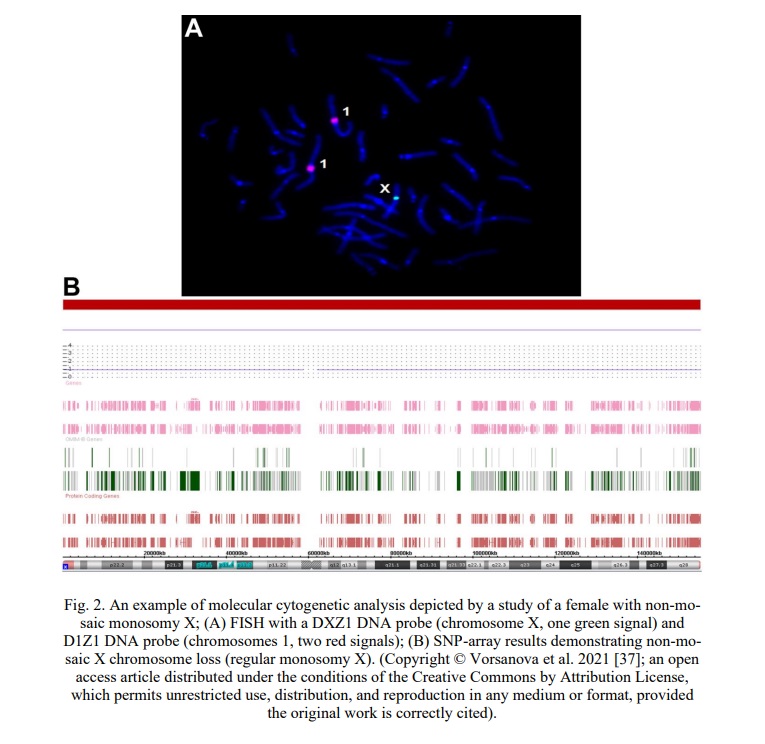
Focusing on karyotyping data obtained from studying 8556 individuals with neurodevelopmental disorders and congenital malformations, allowed us to select a very specific group of individuals (n=600; ~7%), who require molecular karyotyping and extended bioinformatic analysis of genome data. SNP array was selected as a technique of choice, inasmuch as it allows detecting chromosomal imbalances and CNVs at the highest resolution [40-43] and segmental uniparental disomies within imprinted loci [40, 47]. Thus, a heuristic algorithm for comprehensive genome analysis for medical genetics does require both karyotyping (classical cytogenetics) and molecular karyotyping by SNP array. Since FISH allows single-cell monitoring of genomic/chromosomal variations, it is, thereby, required for cases of mosaicism and chromosome/genome instability
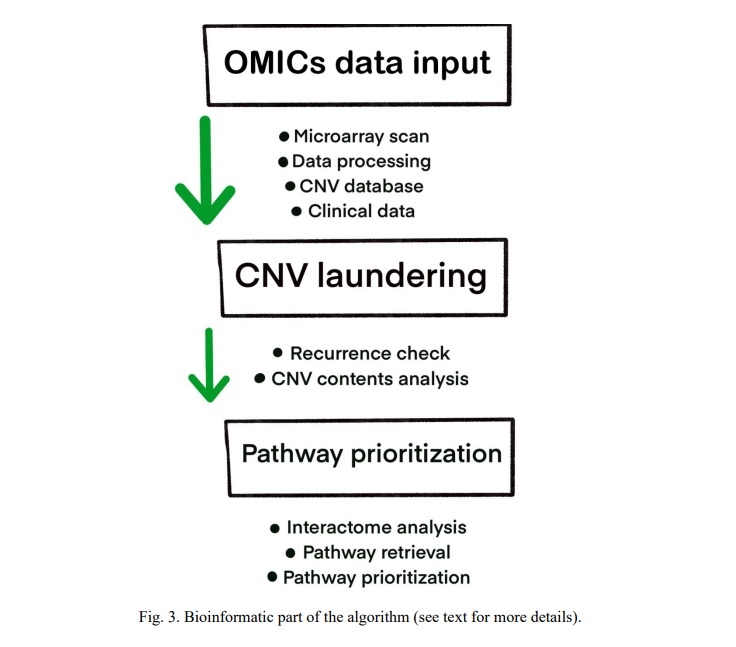
Despite the studies reporting significantly increased diagnostic efficiency rates resulted from the application of array genome scanning techniques, understanding the mechanisms of the diseases and unraveling the cellular and molecular pathways requires a bioinformatic addition to the analytic workflows [16, 28, 32, 36, 44, 48]. Bioinformatic approaches to analyze data obtained by new sequencing technologies including panel versions were suggested to be an alternative [9, 11, 48, 49]. However, the alternative remains questionable, since these approaches do not provide intrinsic data on diseases mechanisms/pathways, inasmuch as genomic mechanisms are much more sophisticated than previously envisaged [20, 28, 50]. Moreover, these mechanisms become even more complicated when somatic genomic mosaicism and chromosome/genome instability are taken into account [3, 7, 15, 25, 51, 52]. Considering the data that applications of the simplest digital tools appreciably increase the yield of molecular cytogenetic genome analysis [1, 17, 53-55], sophisticated bioinformatic techniques are likely to provide the highest efficiency in diagnostic/research cytogenomic analyses [7, 16, 28, 44, 45]. Here, we have added to the algorithm all of our original developments in genomic data processing, which include OMICs data input/processing (variome analysis, data collection, CNV prioritization) [3, 10, 12, 16, 28, 44], CNV laundering [45], pathway-based analyses of variome for candidate process or CNV prioritization [16, 20, 28, 36, 44] (Fig. 3).
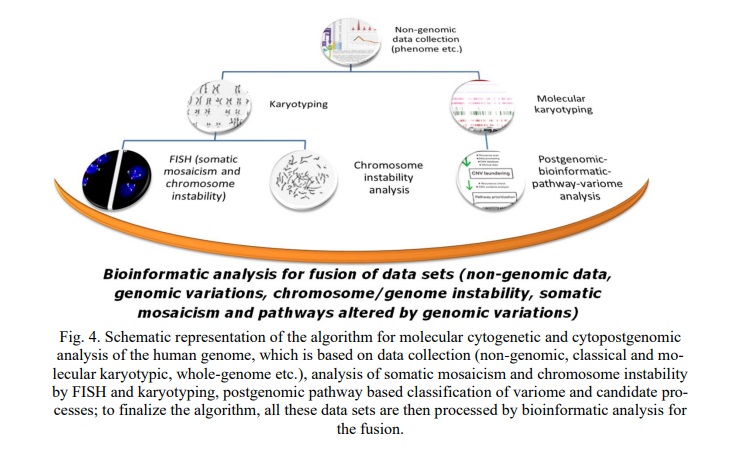
Combining the aforementioned cytogenetic, molecular cytogenetic and cytogenomic or cytopostgenomic approaches seems to result in uncovering mechanisms of a disease associated with a set of genomic variations (disease variome), clinical outcomes of which are mediated by chromosomal/genomic instability and/or molecular and cellular pathways (candidate processes). In this light, it is to note again that morbid conditions associated with chromosomal/genomic instability range from infertility to pathogenic aging, from early onset neurodevelopmental disorders to late onset neurodegenerative and neuropsychiatric diseases, from immunodeficiency to cancer [1-3, 6, 7, 15, 22-25, 29-31, 51, 52]. Finally, the described heuristic algorithm in its final form may be depicted, as follows (Fig. 4).
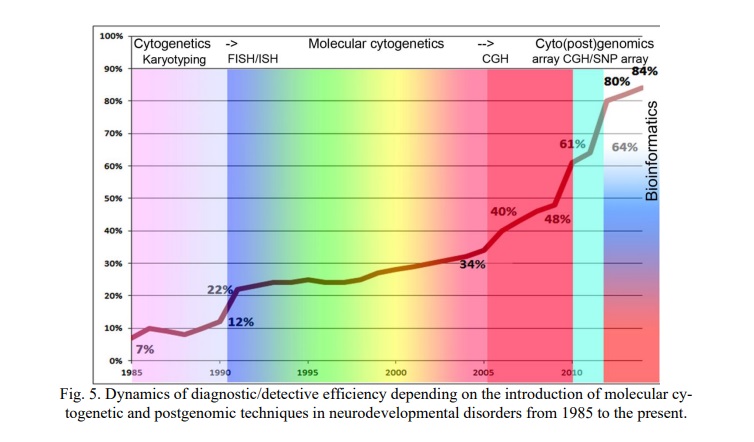
Our long-term experience (from 1985 to the present) [1, 17, 37, 38] allowed us to highlight the dynamics of efficiency changes in dependence of introducing molecular cytogenetic and postgenomic techniques (Fig. 5). Initially, karyotyping uncovers ~7% of genomic variations (chromosomal abnormalities) in neurodevelopmental disorders. The introduction of in situ hybridization (ISH) gave instantly 12% of detection of chromosomal abnormalities. Further introduction and developments of FISH (e.g. increasing DNA probe numbers, multicolor FISH [39], analysis of chromosomal instability and mosaicism) increased the efficiency to 34% (~3 times increase). Metaphase or classical comparative genomic hybridization (CGH) in combination with FISH and karyotyping allowed to detect chromosomal variations in 40% of individuals from the neurodevelopmental cohort. Substitution of CGH by array CGH (CGH on chips; laser detection) increased the efficiency to 48%. SNP array with the highest resolution of molecular karyotyping (up to 1 kbp), used instead of array CGH, gave an unprecedented efficiency in detection of chromosomal abnormalities and CNV (including intragenic CNV) estimated as 61-64% (depending on protocol/variation size filter). Finally, in silico molecular cytogenetic analysis using bioinformatic, postgenomic/cytopostgenomic or pathway-based classification made a breakthrough in efficiency of genomic variation detection, which achieved 80-84%. In this context, we would like to mention that these efficiency rates were achievable by an extremely thorough selection (clinical and cytogenetic selection) of cases to be addressed by genome scanning, FISH and cytopostgenomic methods.
Conclusion. Our communication describes a heuristic algorithm for molecular cytogenetic and cytopostgenomic analysis of the human genome, which may be successfully used to uncover mechanisms for genetic diseases (especially, brain genetic disorders). The algorithm is also applicable for identification of causes and consequences of somatic chromosomal mosaicism and chromosome/genome instability, which are common mechanisms for a wide spectrum of human morbid conditions. The rise of detection rates produced by introducing molecular cytogenetic and cytopostgenomic techniques was a basis for developing the algorithm, the efficiency of which was high enough to conclude that it is highly competitive for research and diagnostic purposes during forthcoming genomic or cyto(post)genomic studies.
Financial support
Our study is partially supported by the Government Assignment of the Russian Ministry of Health, Assignment no. AAAA-A18–118051590122-7 and by the Government Assignment of the Russian Ministry of Science and Higher Education, Assignment no. AAAA-A19–119040490101-6.




















Reference lists